homeobox D3a
ZFIN 
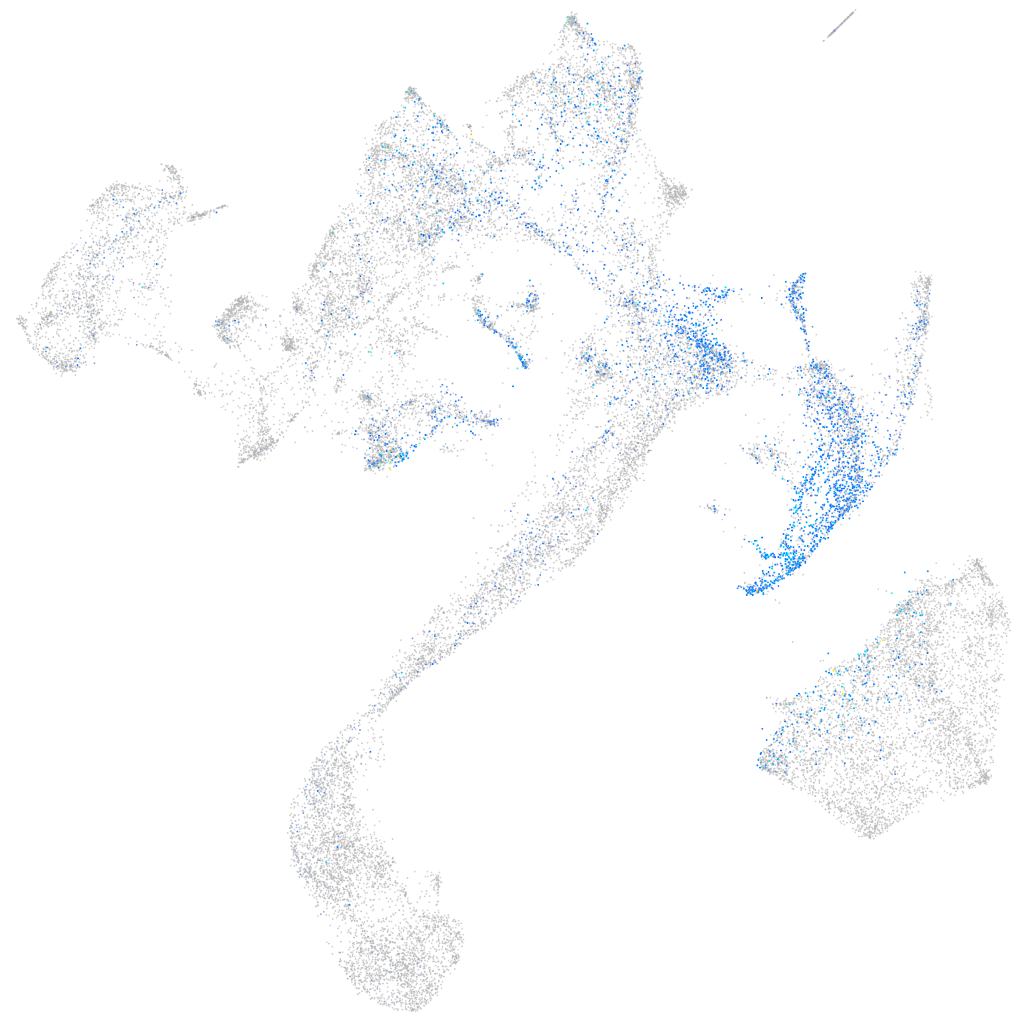
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hoxd10a | 0.372 | actc1b | -0.179 |
| hoxb10a | 0.367 | ttn.2 | -0.168 |
| hoxd12a | 0.329 | atp2a1 | -0.163 |
| hoxa11b | 0.312 | ak1 | -0.162 |
| hoxa10b | 0.311 | tmem38a | -0.161 |
| her12 | 0.310 | aldoab | -0.160 |
| her1 | 0.290 | ckma | -0.160 |
| itm2cb | 0.290 | gapdh | -0.159 |
| hoxa11a | 0.288 | ckmb | -0.158 |
| hoxd11a | 0.281 | pabpc4 | -0.158 |
| sall4 | 0.279 | tnnc2 | -0.154 |
| hoxc11a | 0.268 | neb | -0.149 |
| hoxd9a | 0.268 | mylpfa | -0.149 |
| BX005254.3 | 0.267 | acta1b | -0.148 |
| fbln1 | 0.261 | ttn.1 | -0.147 |
| ephb3 | 0.239 | mybphb | -0.147 |
| hoxc10a | 0.238 | srl | -0.147 |
| nid2a | 0.237 | ldb3a | -0.146 |
| hes6 | 0.235 | klhl41b | -0.145 |
| greb1 | 0.233 | desma | -0.145 |
| gdf11 | 0.232 | eno1a | -0.143 |
| rarga | 0.230 | gamt | -0.143 |
| hnrnpa0l | 0.230 | myl1 | -0.143 |
| hoxc3a | 0.229 | tpma | -0.142 |
| her7 | 0.229 | nme2b.2 | -0.141 |
| h3f3a | 0.226 | si:ch73-367p23.2 | -0.141 |
| hoxb7a | 0.223 | cav3 | -0.139 |
| tspan7 | 0.223 | ldb3b | -0.139 |
| hoxc11b | 0.223 | mylz3 | -0.138 |
| hoxb9a | 0.223 | actn3b | -0.137 |
| thbs2a | 0.219 | cox17 | -0.137 |
| pcdh8 | 0.217 | gatm | -0.137 |
| zbtb16a | 0.214 | pvalb1 | -0.135 |
| LO016987.2 | 0.214 | si:ch211-266g18.10 | -0.135 |
| XLOC-042222 | 0.213 | actn3a | -0.135 |