homeobox D3a
ZFIN 
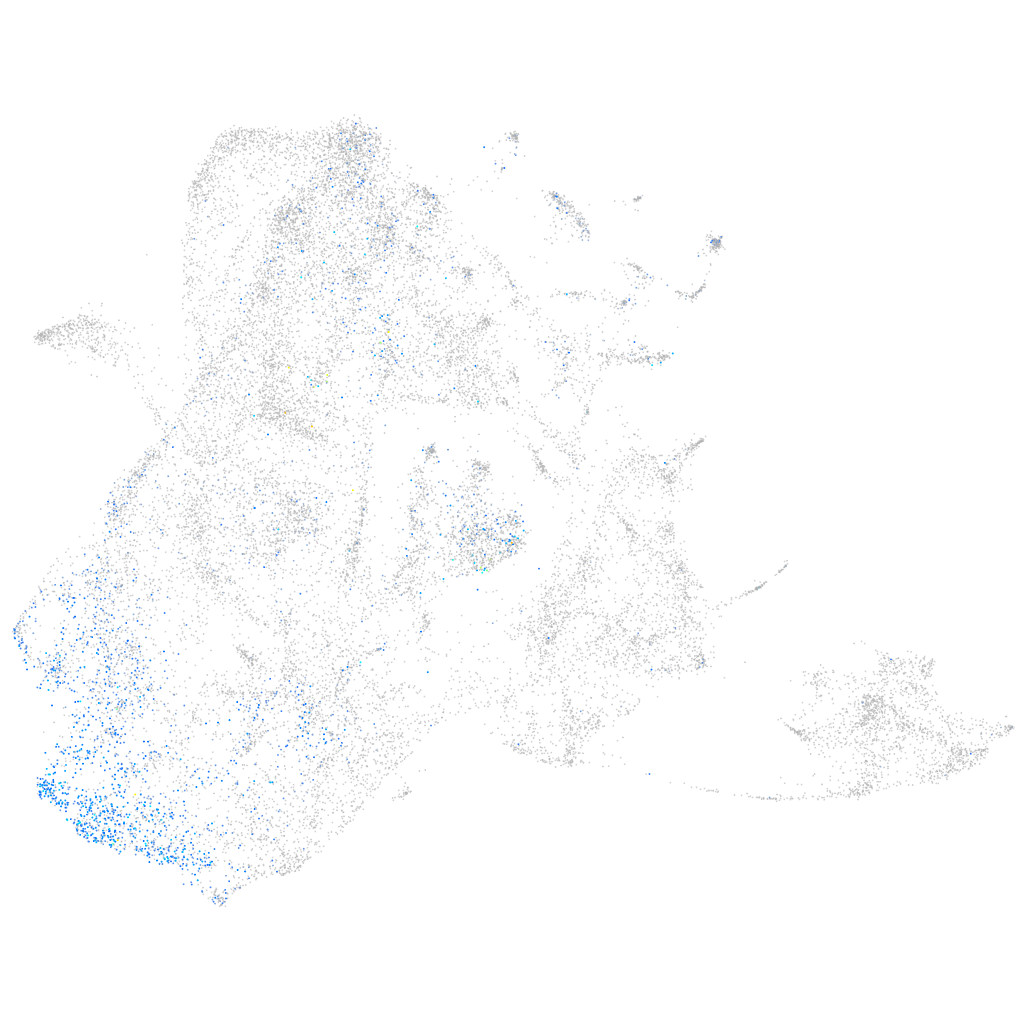
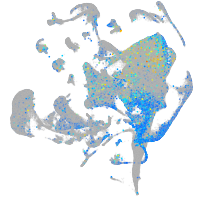
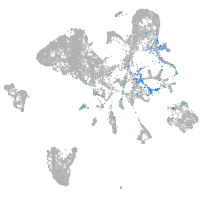
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cdx4 | 0.456 | krt91 | -0.179 |
| hoxb10a | 0.422 | capns1a | -0.171 |
| BX005254.3 | 0.412 | rplp2 | -0.167 |
| hoxd9a | 0.409 | anxa2a | -0.163 |
| hoxc8a | 0.404 | anxa1a | -0.161 |
| hoxb9a | 0.401 | gapdhs | -0.160 |
| hoxb7a | 0.394 | eef1da | -0.156 |
| hoxc3a | 0.391 | ahcy | -0.146 |
| hoxd10a | 0.383 | capgb | -0.146 |
| LOC101882251 | 0.369 | krtt1c19e | -0.143 |
| hoxa10b | 0.366 | rps29 | -0.138 |
| emx2 | 0.362 | aqp3a | -0.136 |
| insl5a | 0.360 | s100v2 | -0.133 |
| vcana | 0.356 | s100v1 | -0.132 |
| apoc1 | 0.355 | si:dkey-33c14.3 | -0.131 |
| hoxc6b | 0.350 | icn | -0.131 |
| sp8b | 0.340 | cxl34b.11 | -0.130 |
| mllt3 | 0.331 | igfbp7 | -0.126 |
| hoxd12a | 0.327 | epgn | -0.126 |
| prickle1a | 0.325 | pvalb1 | -0.121 |
| hoxa9b | 0.317 | pycard | -0.119 |
| crabp2a | 0.313 | pvalb2 | -0.119 |
| hoxd11a | 0.311 | si:ch211-105c13.3 | -0.119 |
| sp8a | 0.306 | col1a1b | -0.117 |
| tmeff1b | 0.298 | cotl1 | -0.116 |
| ccdc3a | 0.298 | ak1 | -0.113 |
| b3gnt7 | 0.296 | caspb | -0.113 |
| hoxa11b | 0.296 | rbp4 | -0.113 |
| nradd | 0.296 | krt5 | -0.111 |
| tpm4a | 0.288 | gstm.3 | -0.110 |
| hoxc11b | 0.287 | zgc:136930 | -0.109 |
| hoxa9a | 0.284 | si:ch211-39i22.1 | -0.109 |
| cdaa | 0.282 | col1a1a | -0.109 |
| ptgs1 | 0.274 | s100a10b | -0.107 |
| mdka | 0.273 | fbp1a | -0.106 |