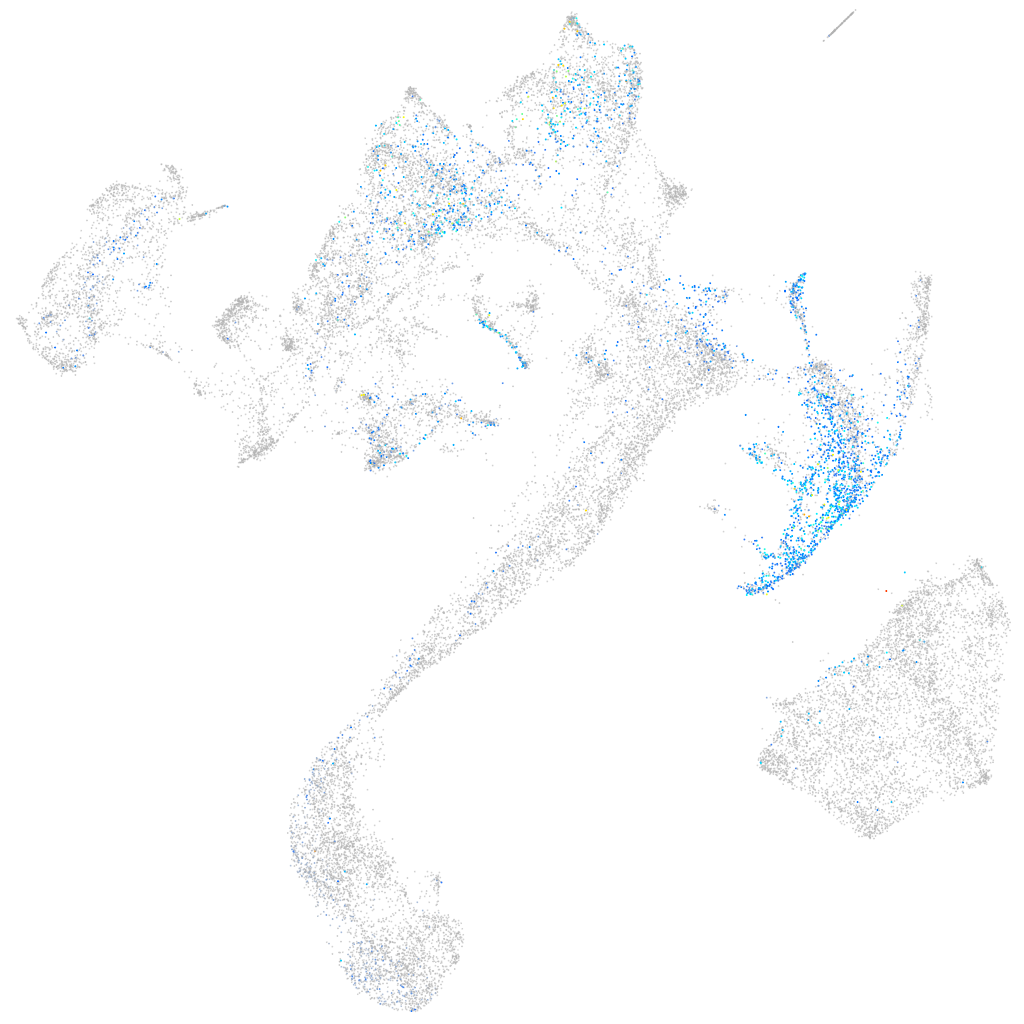
homeobox C11a
ZFIN 
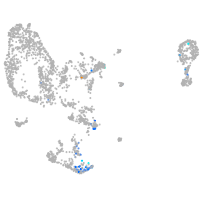
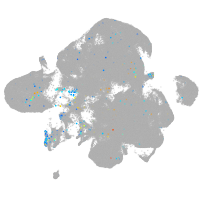
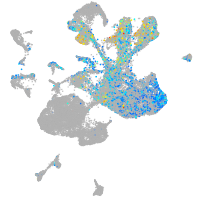
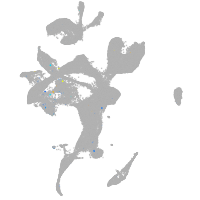
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hoxd12a | 0.469 | ttn.2 | -0.157 |
| hoxd10a | 0.442 | ttn.1 | -0.147 |
| hoxc11b | 0.395 | actc1b | -0.143 |
| hoxb10a | 0.389 | klhl41b | -0.134 |
| hoxa11a | 0.383 | tmem38a | -0.134 |
| hoxa11b | 0.350 | aldoab | -0.133 |
| hoxd11a | 0.349 | fxr2 | -0.132 |
| her1 | 0.348 | ak1 | -0.130 |
| itm2cb | 0.346 | pabpc4 | -0.129 |
| hoxa10b | 0.336 | gapdh | -0.128 |
| her12 | 0.320 | atp2a1 | -0.127 |
| hoxc10a | 0.314 | desma | -0.127 |
| her7 | 0.292 | srl | -0.127 |
| ephb3 | 0.281 | tnni2b.1 | -0.124 |
| hoxd9a | 0.274 | cox7c | -0.124 |
| sall4 | 0.270 | ldb3a | -0.124 |
| hoxd3a | 0.268 | mybphb | -0.123 |
| greb1 | 0.264 | tnnc2 | -0.123 |
| BX005254.3 | 0.261 | ckma | -0.123 |
| fbln1 | 0.259 | zgc:101853 | -0.123 |
| rarga | 0.256 | ckmb | -0.122 |
| pcdh8 | 0.247 | acta1b | -0.120 |
| hes6 | 0.244 | eno1a | -0.120 |
| tbx16l | 0.244 | acta1a | -0.120 |
| msgn1 | 0.240 | cav3 | -0.119 |
| LO016987.2 | 0.240 | rbfox1l | -0.119 |
| gdf11 | 0.235 | tpma | -0.118 |
| thbs2a | 0.233 | txlnbb | -0.118 |
| zgc:162939 | 0.220 | neb | -0.118 |
| hnrnpa0l | 0.220 | nme2b.2 | -0.118 |
| nid2a | 0.217 | gatm | -0.117 |
| zbtb16a | 0.216 | cox17 | -0.117 |
| h3f3a | 0.215 | zgc:92518 | -0.117 |
| phgdh | 0.210 | myl1 | -0.116 |
| fzd10 | 0.210 | CABZ01072309.1 | -0.115 |