homeobox D10a
ZFIN 
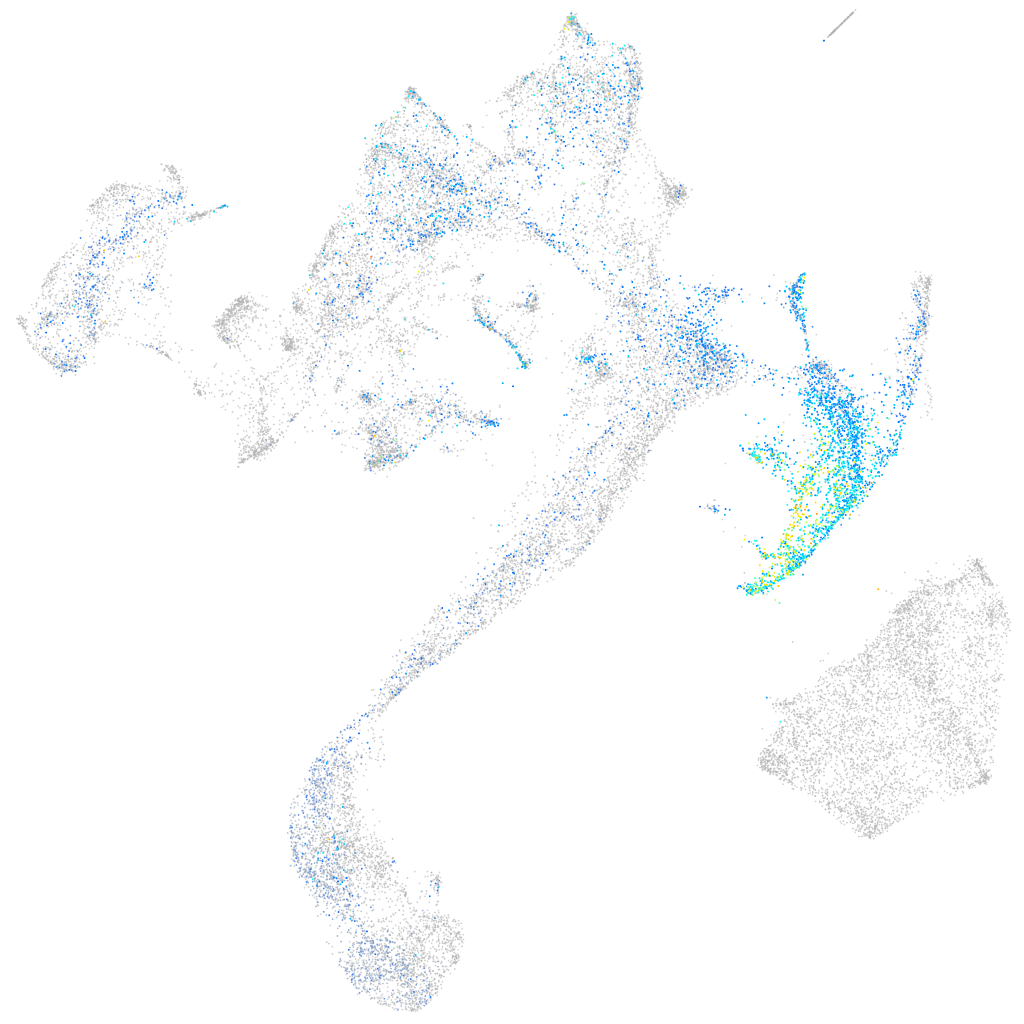
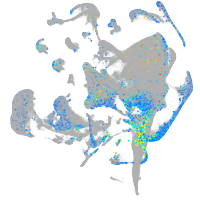
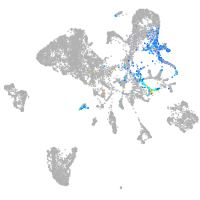
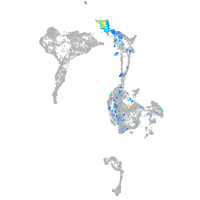
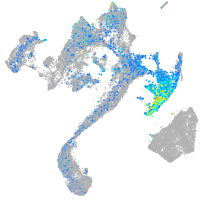
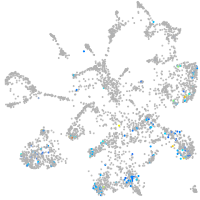
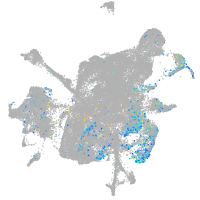
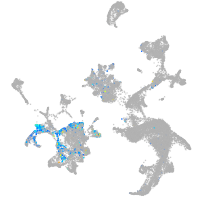
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hoxb10a | 0.634 | actc1b | -0.182 |
| her1 | 0.555 | ttn.2 | -0.179 |
| hoxd12a | 0.531 | pabpc4 | -0.161 |
| itm2cb | 0.511 | tmem38a | -0.159 |
| her12 | 0.503 | klhl41b | -0.157 |
| hoxa11b | 0.478 | akap12b | -0.155 |
| hoxa10b | 0.478 | si:ch211-152c2.3 | -0.154 |
| hoxd11a | 0.459 | ak1 | -0.154 |
| BX005254.3 | 0.452 | fabp3 | -0.153 |
| hoxa11a | 0.451 | atp2a1 | -0.153 |
| hoxc11a | 0.442 | cox7c | -0.152 |
| her7 | 0.433 | eno1a | -0.151 |
| sall4 | 0.405 | rps18 | -0.151 |
| pcdh8 | 0.405 | ckma | -0.150 |
| ephb3 | 0.402 | ttn.1 | -0.150 |
| hes6 | 0.391 | aldoab | -0.149 |
| hoxd9a | 0.390 | ckmb | -0.147 |
| hoxc11b | 0.385 | mylpfa | -0.147 |
| greb1 | 0.374 | gapdh | -0.147 |
| hoxd3a | 0.372 | desma | -0.147 |
| msgn1 | 0.366 | srl | -0.144 |
| nid2a | 0.357 | NC-002333.4 | -0.143 |
| LO016987.2 | 0.356 | zgc:101853 | -0.143 |
| tbx16l | 0.350 | gatm | -0.142 |
| hoxc10a | 0.350 | fxr2 | -0.142 |
| hnrnpa0l | 0.345 | tnnc2 | -0.142 |
| hoxb7a | 0.328 | FQ323156.1 | -0.141 |
| apoc1 | 0.325 | txlnbb | -0.141 |
| rarga | 0.324 | gamt | -0.140 |
| fbln1 | 0.323 | sparc | -0.140 |
| thbs2a | 0.320 | stm | -0.139 |
| h3f3a | 0.315 | myl1 | -0.139 |
| gdf11 | 0.312 | cox17 | -0.138 |
| tspan7 | 0.309 | cav3 | -0.138 |
| zgc:162939 | 0.304 | ldb3a | -0.138 |