homeobox B10a
ZFIN 
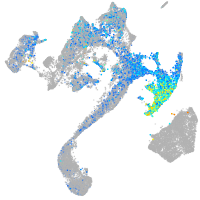
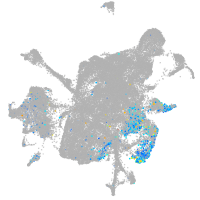
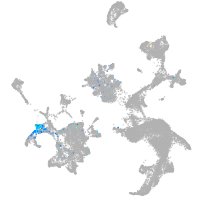
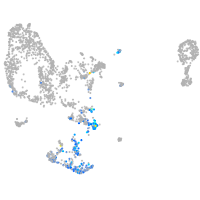
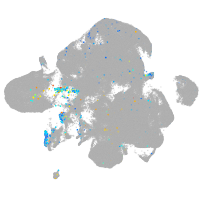
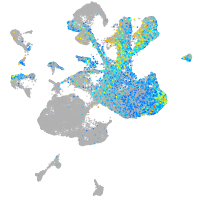
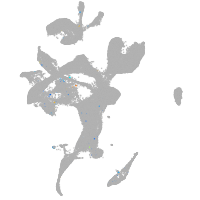
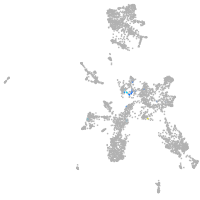
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| AL954146.1 | 0.435 | anxa5b | -0.056 |
| si:dkey-191j3.2 | 0.384 | mif | -0.051 |
| hoxd13a | 0.364 | capns1b | -0.050 |
| mnx2b | 0.322 | cst14a.2 | -0.049 |
| hoxd9a | 0.321 | anxa11b | -0.048 |
| mab21l3 | 0.317 | gsto2 | -0.046 |
| LOC108190371 | 0.299 | calm1b | -0.045 |
| hoxd12a | 0.292 | mgst3b | -0.045 |
| hoxb9a | 0.290 | mcl1b | -0.044 |
| hoxc12a | 0.289 | anxa1a | -0.044 |
| hoxc11a | 0.269 | zbtb10 | -0.043 |
| CR376751.1 | 0.269 | mhc1zba | -0.043 |
| hoxc12b | 0.257 | tspan15 | -0.043 |
| hoxc6b | 0.257 | atp1b1a | -0.043 |
| si:dkey-36i7.3 | 0.252 | cdh1 | -0.042 |
| pcdh1g1 | 0.251 | ppa1b | -0.042 |
| hoxa10b | 0.251 | nqo1 | -0.042 |
| LOC108190059.1 | 0.247 | mt-co2 | -0.041 |
| hamp | 0.246 | mt-cyb | -0.041 |
| CABZ01059408.1 | 0.240 | naga | -0.040 |
| pcdh2ab1 | 0.239 | ucp2 | -0.040 |
| CT573337.1 | 0.236 | zgc:165573 | -0.040 |
| slc18a3a | 0.235 | selenow1 | -0.040 |
| mxra5a | 0.235 | atp1a1a.1 | -0.040 |
| zgc:112334 | 0.233 | zgc:158343 | -0.040 |
| BX908753.1 | 0.223 | zgc:101744 | -0.039 |
| rnf220b | 0.222 | cox7a2l | -0.039 |
| tmie | 0.219 | ist1 | -0.039 |
| sema3e | 0.210 | s100v2 | -0.039 |
| txnipb | 0.209 | gstp1 | -0.039 |
| hoxc8a | 0.208 | prelid3b | -0.039 |
| ebi3 | 0.203 | higd1a | -0.039 |
| chata | 0.202 | selenow2b | -0.038 |
| neurod2 | 0.200 | zgc:113333 | -0.038 |
| scrt2 | 0.197 | sult2st1 | -0.038 |