motor neuron and pancreas homeobox 2b
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 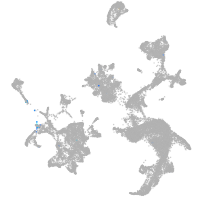 |
pronephros  |
neural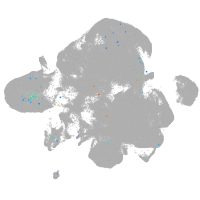 |
spinal cord / glia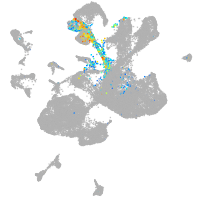 |
eye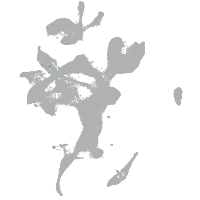 |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mnx2a | 0.335 | celf2 | -0.032 |
| mnx1 | 0.300 | eef1da | -0.029 |
| slc18a3a | 0.236 | ahnak | -0.027 |
| lhx4 | 0.205 | sparc | -0.027 |
| chata | 0.198 | anxa1a | -0.026 |
| si:ch211-247j9.1 | 0.198 | cfl1l | -0.026 |
| slc5a7a | 0.183 | cyt1 | -0.026 |
| LOC100537369 | 0.181 | krt5 | -0.025 |
| sv2c | 0.163 | s100a10b | -0.025 |
| tac1 | 0.140 | krtt1c19e | -0.024 |
| ret | 0.135 | s100v2 | -0.024 |
| isl2a | 0.134 | anxa2a | -0.023 |
| slit3 | 0.127 | ckap4 | -0.023 |
| alcama | 0.123 | cyt1l | -0.023 |
| prph | 0.122 | si:dkey-16p21.8 | -0.023 |
| chodl | 0.120 | cebpd | -0.022 |
| gfra3 | 0.119 | si:ch211-195b11.3 | -0.022 |
| sox32 | 0.118 | zgc:162730 | -0.022 |
| hoxb9a | 0.117 | fxyd6l | -0.021 |
| sox17 | 0.114 | gsnb | -0.021 |
| LHX3 | 0.113 | hrc | -0.021 |
| hoxa9a | 0.109 | icn | -0.021 |
| hoxa9b | 0.106 | icn2 | -0.021 |
| tuba8l3 | 0.105 | ier2b | -0.021 |
| vim | 0.105 | krt17 | -0.021 |
| si:dkey-178e17.1 | 0.104 | mab21l1 | -0.021 |
| tmie | 0.103 | si:ch211-105c13.3 | -0.021 |
| hoxd9a | 0.099 | slc38a5b | -0.021 |
| isl1 | 0.098 | slc6a1b | -0.021 |
| kcnf1a | 0.095 | anxa1c | -0.020 |
| myt1b | 0.094 | cd99 | -0.020 |
| ptprfa | 0.092 | cldne | -0.020 |
| XLOC-007078 | 0.092 | col1a2 | -0.020 |
| gdf11 | 0.087 | epcam | -0.020 |
| SLC35D3 | 0.087 | pfn1 | -0.020 |




