motor neuron and pancreas homeobox 2b
ZFIN 
Other cell groups


all cells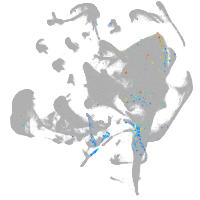 |
blastomeres |
PGCs |
endoderm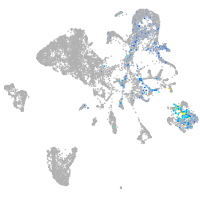 |
axial mesoderm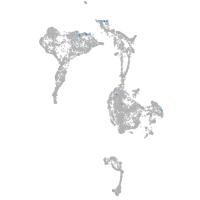 |
muscle 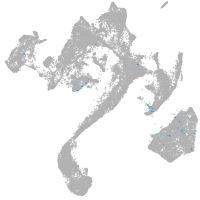 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 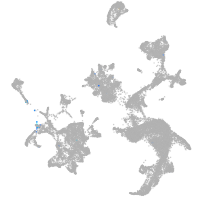 |
pronephros  |
neural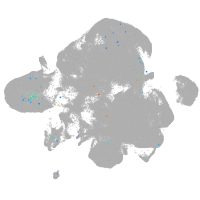 |
spinal cord / glia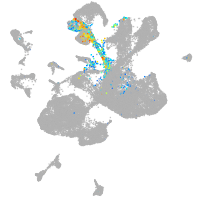 |
eye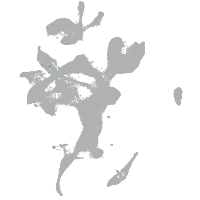 |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mnx1 | 0.266 | rps28 | -0.030 |
| chata | 0.206 | uba52 | -0.028 |
| slc18a3a | 0.163 | rpl35 | -0.027 |
| si:ch211-217i17.1 | 0.141 | rps8a | -0.027 |
| or113-1 | 0.136 | rplp1 | -0.027 |
| CABZ01025311.1 | 0.131 | eef1b2 | -0.026 |
| LOC108190371 | 0.117 | pmp22a | -0.026 |
| XLOC-011873 | 0.110 | rps24 | -0.024 |
| LOC101885930 | 0.106 | rpl9 | -0.024 |
| cntn4 | 0.104 | rpl31 | -0.022 |
| mnx2a | 0.103 | rpl12 | -0.022 |
| uts2d | 0.103 | rpl24 | -0.021 |
| slc5a7a | 0.102 | rpl23 | -0.021 |
| esrrga | 0.102 | rpl39 | -0.021 |
| sv2c | 0.098 | rps3a | -0.021 |
| card14 | 0.097 | rps15 | -0.021 |
| BX276128.1 | 0.097 | sparc | -0.020 |
| tac1 | 0.096 | rps29 | -0.020 |
| prph | 0.095 | rps26l | -0.020 |
| mir725 | 0.094 | col1a2 | -0.020 |
| pimr138 | 0.093 | rps15a | -0.020 |
| baiap2l2a | 0.091 | mfap2 | -0.019 |
| CR936416.1 | 0.090 | rack1 | -0.019 |
| lhx4 | 0.090 | rpl18a | -0.019 |
| csf2rb | 0.089 | rpl23a | -0.019 |
| LOC108192140 | 0.088 | rps27a | -0.019 |
| acot17 | 0.087 | rpl8 | -0.019 |
| LOC100537369 | 0.085 | rpl22 | -0.019 |
| myt1b | 0.085 | rpl30 | -0.019 |
| l1cama | 0.085 | eef1da | -0.018 |
| si:ch211-247j9.1 | 0.084 | si:ch211-286o17.1 | -0.017 |
| CABZ01080371.3 | 0.082 | rps9 | -0.017 |
| CR392001.2 | 0.080 | rpl26 | -0.017 |
| zgc:153142 | 0.078 | rpl14 | -0.017 |
| slc18a3b | 0.077 | rpl7a | -0.016 |




