BAR/IMD domain containing adaptor protein 2 like 2a
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 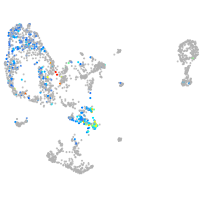 |
neural |
spinal cord / glia |
eye |
taste / olfactory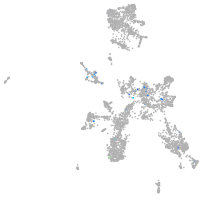 |
otic / lateral line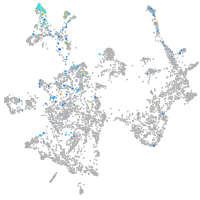 |
epidermis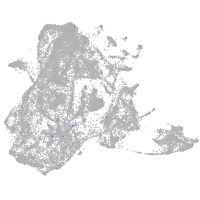 |
periderm |
fin |
ionocytes / mucous |
pigment cells |
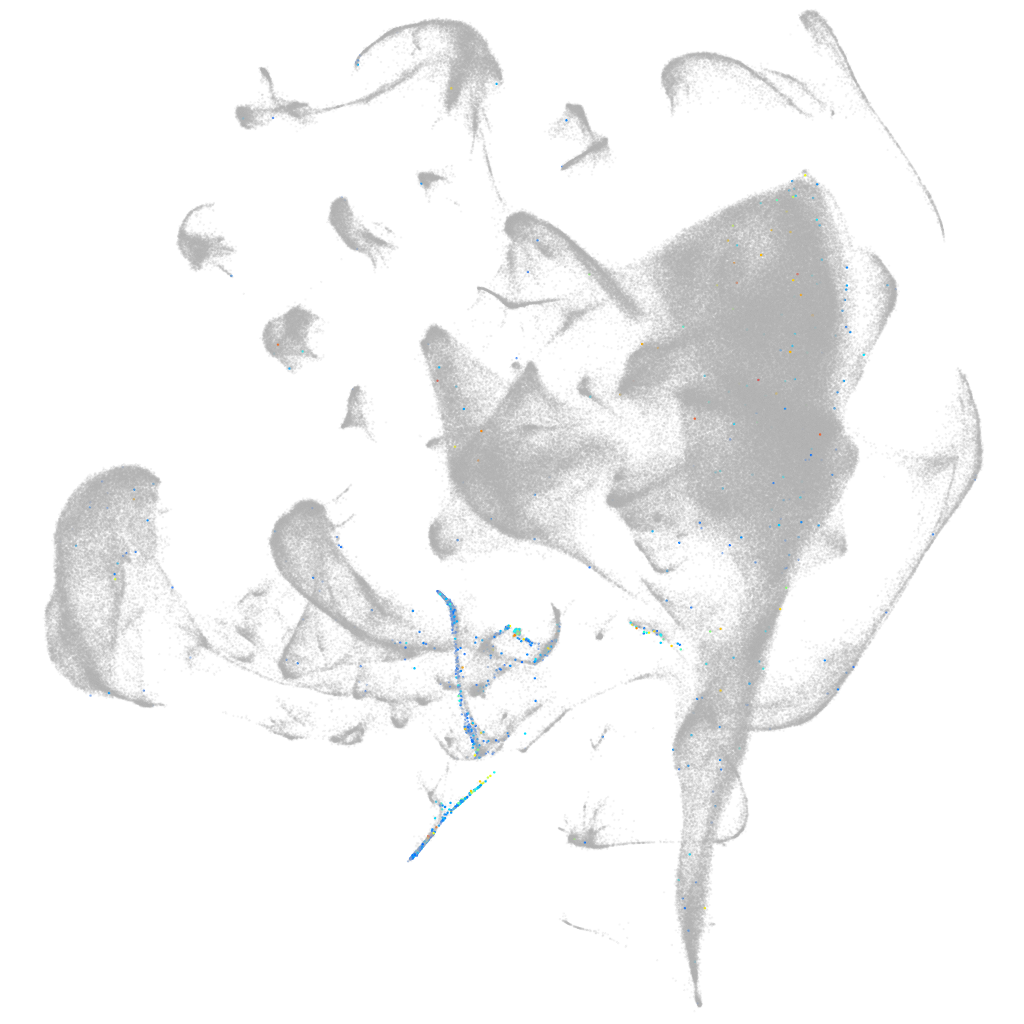
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cdh17 | 0.278 | fabp3 | -0.040 |
| pdzk1 | 0.278 | marcksl1a | -0.038 |
| lrrc66 | 0.256 | gpm6aa | -0.037 |
| eps8l3b | 0.255 | gpm6ab | -0.035 |
| anpepb | 0.251 | tuba1c | -0.035 |
| IZUMO1R | 0.250 | celf2 | -0.034 |
| sdsl | 0.244 | nova2 | -0.033 |
| atp1a1a.4 | 0.243 | mdkb | -0.032 |
| eps8l3a | 0.242 | rtn1a | -0.032 |
| ezra | 0.242 | mdka | -0.030 |
| mbl2 | 0.241 | tubb5 | -0.030 |
| LOC100536156 | 0.240 | actc1b | -0.029 |
| muc13b | 0.240 | cadm3 | -0.029 |
| slc6a19a.2 | 0.240 | atp6v0cb | -0.028 |
| slc6a19b | 0.239 | stmn1b | -0.028 |
| si:dkey-30c15.13 | 0.233 | cspg5a | -0.027 |
| si:ch211-133l5.7 | 0.232 | elavl3 | -0.026 |
| enpep | 0.231 | fabp7a | -0.026 |
| ggt1b | 0.230 | rtn1b | -0.026 |
| si:ch73-185c24.2 | 0.230 | hmgb1b | -0.025 |
| CU682777.2 | 0.227 | atp1a3a | -0.024 |
| pls1 | 0.227 | dbn1 | -0.024 |
| epdl2 | 0.224 | sypb | -0.024 |
| slc6a19a.1 | 0.221 | fam168a | -0.023 |
| calml4a | 0.220 | gng3 | -0.023 |
| AL831745.1 | 0.219 | pvalb1 | -0.023 |
| hnf4g | 0.218 | pvalb2 | -0.023 |
| zanl | 0.218 | si:ch211-137a8.4 | -0.023 |
| dpp4 | 0.217 | atp1b2a | -0.022 |
| clic5b | 0.217 | epb41a | -0.022 |
| LOC103908636 | 0.216 | hmgb3a | -0.022 |
| si:dkey-194e6.1 | 0.216 | meis1b | -0.022 |
| si:ch211-117n7.7 | 0.215 | mylpfa | -0.022 |
| myo15b | 0.214 | nat8l | -0.022 |
| bin2a | 0.212 | si:dkeyp-75h12.5 | -0.022 |