BAR/IMD domain containing adaptor protein 2 like 2a
ZFIN 
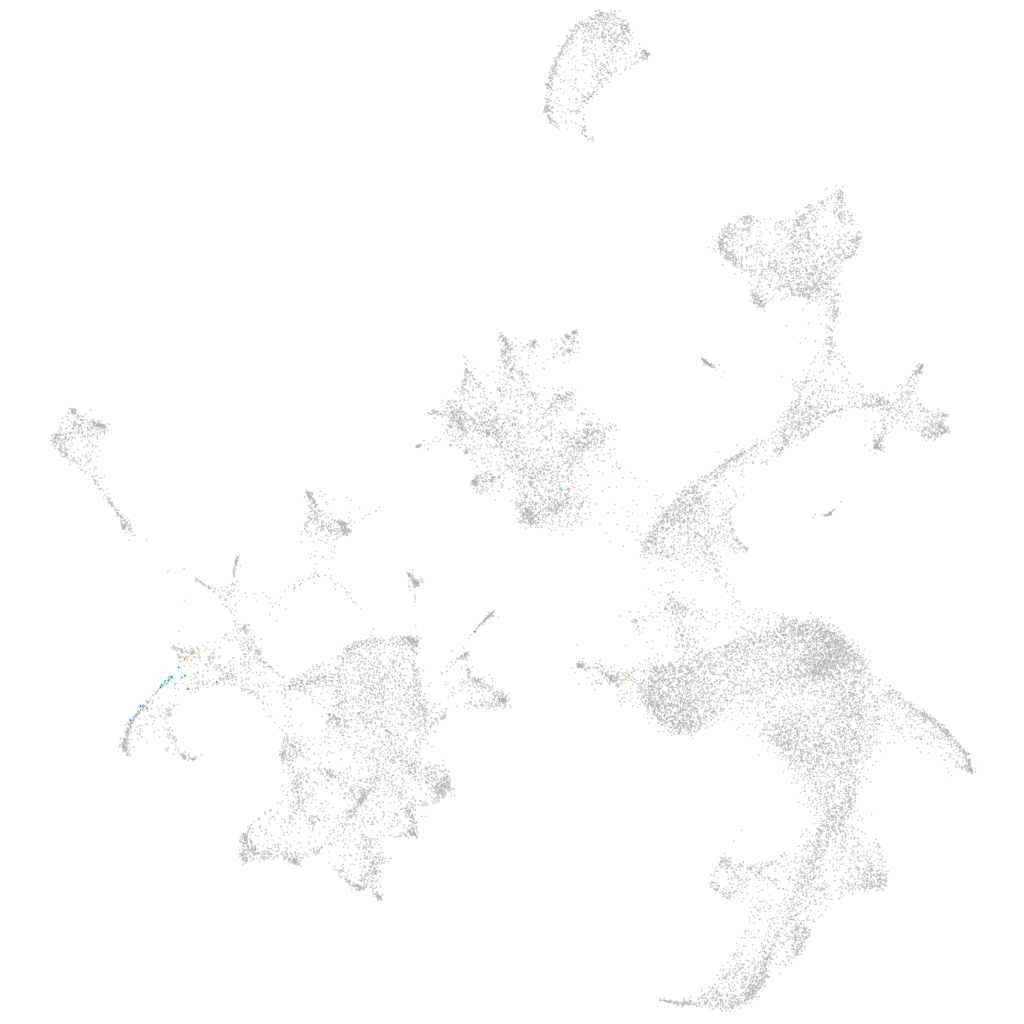
Other cell groups


all cells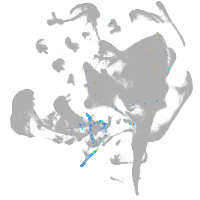 |
blastomeres |
PGCs |
endoderm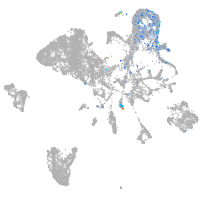 |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 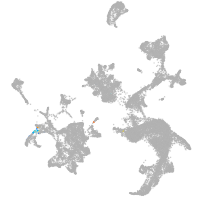 |
pronephros 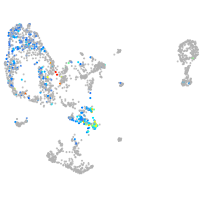 |
neural |
spinal cord / glia |
eye |
taste / olfactory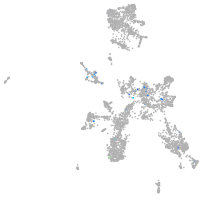 |
otic / lateral line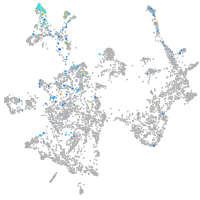 |
epidermis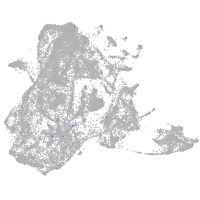 |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC101884976 | 0.350 | hbbe1.3 | -0.030 |
| mapk15 | 0.285 | hbae1.1 | -0.028 |
| si:ch211-220f16.1 | 0.268 | hbae3 | -0.028 |
| ttc12 | 0.257 | hbae1.3 | -0.027 |
| XLOC-042792 | 0.252 | hbbe1.1 | -0.026 |
| LOC101885806 | 0.252 | hemgn | -0.024 |
| daw1 | 0.232 | hbbe2 | -0.024 |
| ccdc173 | 0.213 | hbbe1.2 | -0.024 |
| LOC103910744.1 | 0.205 | cahz | -0.024 |
| fbxo16 | 0.204 | blvrb | -0.023 |
| prdm6 | 0.194 | slc4a1a | -0.023 |
| ccdc151 | 0.187 | prdx2 | -0.022 |
| pimr28 | 0.185 | tspo | -0.021 |
| foxj1a | 0.176 | zgc:163057 | -0.021 |
| ribc2 | 0.172 | alas2 | -0.019 |
| map3k19 | 0.165 | nmt1b | -0.019 |
| efcab2 | 0.164 | creg1 | -0.019 |
| cldnc | 0.162 | fth1a | -0.018 |
| tekt1 | 0.161 | si:ch211-250g4.3 | -0.018 |
| spag6 | 0.160 | si:ch211-227m13.1 | -0.018 |
| si:ch211-163l21.7 | 0.155 | nt5c2l1 | -0.017 |
| LOC110438469 | 0.155 | mylpfa | -0.017 |
| vcana | 0.153 | epb41b | -0.016 |
| ccdc114 | 0.152 | pvalb2 | -0.016 |
| morc3b | 0.149 | tmem14ca | -0.016 |
| mnx1 | 0.148 | gpx1a | -0.016 |
| tspan35 | 0.148 | eef1da | -0.016 |
| zgc:171699 | 0.146 | sptb | -0.015 |
| ccdc65 | 0.144 | bnip3lb | -0.015 |
| drc1 | 0.142 | crema | -0.015 |
| tex36 | 0.141 | plac8l1 | -0.015 |
| morn3 | 0.140 | klf1 | -0.015 |
| dnaaf4 | 0.139 | zgc:163080 | -0.015 |
| cldn7b | 0.138 | mibp2 | -0.015 |
| LOC110438065 | 0.137 | sod1 | -0.014 |