"Pim proto-oncogene, serine/threonine kinase, related 138"
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 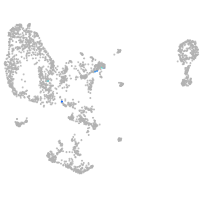 |
neural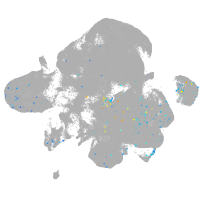 |
spinal cord / glia |
eye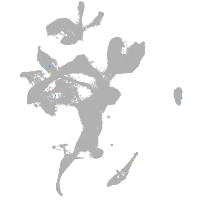 |
taste / olfactory |
otic / lateral line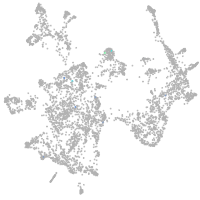 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tmsb | 0.090 | eef1da | -0.025 |
| inab | 0.088 | fosab | -0.025 |
| onecut1 | 0.084 | COX7A2 (1 of many) | -0.024 |
| gap43 | 0.082 | junba | -0.023 |
| mllt11 | 0.081 | mdka | -0.023 |
| ppp1r14ba | 0.074 | tuba8l | -0.023 |
| tubb5 | 0.072 | zgc:162730 | -0.023 |
| pou2f2a | 0.072 | ahnak | -0.022 |
| stmn1b | 0.071 | btg2 | -0.022 |
| map1b | 0.070 | cd63 | -0.022 |
| gng3 | 0.069 | ckap4 | -0.022 |
| LOC100537384 | 0.069 | krt5 | -0.022 |
| dpysl3 | 0.068 | sdc4 | -0.022 |
| elavl3 | 0.068 | sparc | -0.022 |
| stmn2a | 0.064 | tuba8l2 | -0.022 |
| tmeff1b | 0.063 | cyt1 | -0.021 |
| gng2 | 0.062 | hdlbpa | -0.021 |
| si:dkey-276j7.1 | 0.062 | krt4 | -0.021 |
| zc4h2 | 0.062 | pfn1 | -0.021 |
| bsx | 0.061 | s100a10b | -0.021 |
| cxxc4 | 0.061 | si:ch211-286b5.5 | -0.021 |
| rtn1b | 0.061 | si:dkey-16p21.8 | -0.021 |
| elavl4 | 0.060 | anxa1a | -0.020 |
| maptb | 0.060 | ccng1 | -0.020 |
| rtn1a | 0.059 | cebpd | -0.020 |
| si:dkey-56m19.5 | 0.058 | cst14b.1 | -0.020 |
| ywhah | 0.058 | cyt1l | -0.020 |
| celf3a | 0.056 | GCA | -0.020 |
| fez1 | 0.056 | pnp5a | -0.020 |
| id4 | 0.056 | pycard | -0.020 |
| si:dkey-280e21.3 | 0.056 | tspo | -0.020 |
| dpysl2b | 0.056 | XLOC-044191 | -0.020 |
| stmn2b | 0.055 | zgc:153284 | -0.020 |
| tuba1a | 0.055 | zgc:56493 | -0.020 |
| fscn1a | 0.054 | anxa2a | -0.019 |




