"Pim proto-oncogene, serine/threonine kinase, related 138"
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 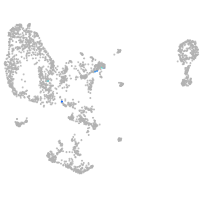 |
neural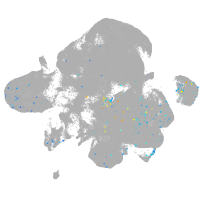 |
spinal cord / glia |
eye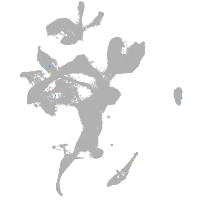 |
taste / olfactory |
otic / lateral line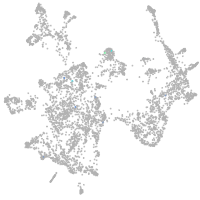 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| taar20q | 0.153 | id1 | -0.014 |
| taar10a | 0.153 | tpm4a | -0.014 |
| taar11 | 0.153 | pmp22a | -0.013 |
| si:ch211-217i17.1 | 0.150 | tuba8l | -0.013 |
| syt18b | 0.146 | fstl1b | -0.013 |
| mir725 | 0.100 | anp32b | -0.012 |
| si:dkeyp-82a1.8 | 0.097 | marveld1 | -0.012 |
| XLOC-034224 | 0.093 | fkbp7 | -0.012 |
| mnx2b | 0.093 | cxcl18b | -0.011 |
| acot17 | 0.092 | dut | -0.011 |
| CABZ01080371.3 | 0.088 | selenoh | -0.011 |
| LOC101885749 | 0.087 | boc | -0.011 |
| cysltr3 | 0.086 | angptl4 | -0.011 |
| npy1r | 0.083 | crtap | -0.011 |
| CR450808.2 | 0.083 | si:dkey-261h17.1 | -0.011 |
| zgc:153142 | 0.083 | col16a1 | -0.011 |
| cd4-2.2 | 0.080 | cnn2 | -0.011 |
| LOC110437727 | 0.076 | rpl27a | -0.011 |
| insm1b | 0.074 | pcna | -0.010 |
| tmsb | 0.072 | rps29 | -0.010 |
| BX539313.1 | 0.071 | dusp1 | -0.010 |
| chata | 0.070 | lig1 | -0.010 |
| rln3b | 0.069 | emd | -0.010 |
| plpp4 | 0.069 | cyyr1 | -0.010 |
| stmn1b | 0.069 | spry2 | -0.010 |
| si:dkey-208c12.2 | 0.068 | ptbp1b | -0.010 |
| LOC110439805 | 0.068 | mfap2 | -0.010 |
| LOC101884125 | 0.067 | fzd7b | -0.010 |
| elavl3 | 0.066 | sdr16c5b | -0.010 |
| mgat3a | 0.065 | slbp | -0.010 |
| insm1a | 0.065 | ppfibp1a | -0.010 |
| si:ch211-191a16.2 | 0.065 | rspo3 | -0.010 |
| si:ch73-261i21.5 | 0.064 | prim2 | -0.010 |
| si:ch1073-153i20.2.1 | 0.061 | snai1a | -0.010 |
| cxcr3.1 | 0.061 | cenpx | -0.010 |




