Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural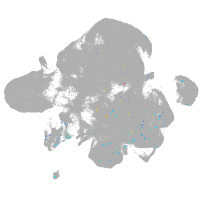 |
spinal cord / glia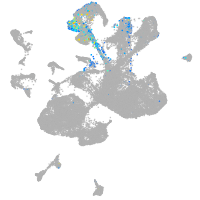 |
eye |
taste / olfactory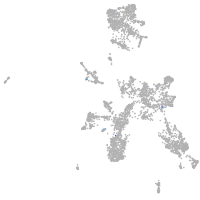 |
otic / lateral line |
epidermis |
periderm |
fin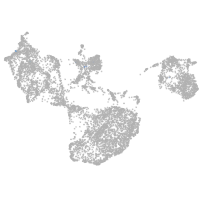 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slc18a3a | 0.260 | aldob | -0.038 |
| slc5a7a | 0.208 | hspb1 | -0.033 |
| chata | 0.198 | atp1b1a | -0.031 |
| mnx1 | 0.194 | eif4ebp3l | -0.031 |
| si:ch211-247j9.1 | 0.190 | banf1 | -0.030 |
| mnx2b | 0.181 | bzw1b | -0.030 |
| tac1 | 0.172 | dkc1 | -0.030 |
| mnx2a | 0.163 | epcam | -0.030 |
| sv2c | 0.159 | id1 | -0.030 |
| chodl | 0.154 | nop58 | -0.030 |
| isl2a | 0.148 | si:ch211-152c2.3 | -0.030 |
| nefmb | 0.143 | apoeb | -0.029 |
| prph | 0.143 | f11r.1 | -0.029 |
| chrnb4 | 0.136 | fbl | -0.029 |
| lhx4 | 0.134 | hdlbpa | -0.029 |
| cplx2l | 0.123 | sdc4 | -0.029 |
| slit3 | 0.121 | stm | -0.029 |
| tmie | 0.119 | vamp3 | -0.029 |
| si:dkey-178e17.1 | 0.117 | ccnd1 | -0.028 |
| ache | 0.116 | chaf1a | -0.028 |
| ret | 0.116 | eef1da | -0.028 |
| si:dkey-85n7.6 | 0.112 | lig1 | -0.028 |
| cplx2 | 0.111 | mki67 | -0.028 |
| arhgap22 | 0.109 | ucp2 | -0.028 |
| SLC35D3 | 0.109 | apoc1 | -0.027 |
| gfra3 | 0.108 | ccng1 | -0.027 |
| neflb | 0.108 | cfl1l | -0.027 |
| pcsk1nl | 0.107 | npm1a | -0.027 |
| syt2a | 0.107 | polr3gla | -0.027 |
| stxbp1a | 0.105 | pou5f3 | -0.027 |
| XLOC-016165 | 0.104 | arf1 | -0.026 |
| snap25a | 0.103 | cast | -0.026 |
| onecut1 | 0.103 | cdh1 | -0.026 |
| chga | 0.102 | nop56 | -0.026 |
| onecut3a | 0.102 | rsl1d1 | -0.026 |




