one cut homeobox 3a
ZFIN 
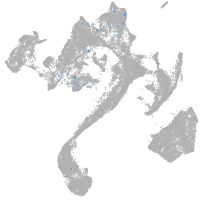
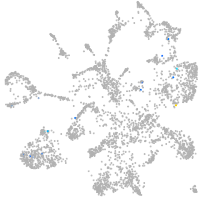
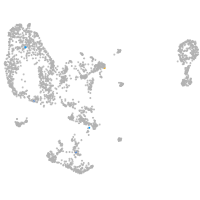
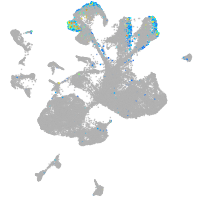
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| XLOC-016165 | 0.365 | id1 | -0.061 |
| onecut1 | 0.320 | vamp3 | -0.057 |
| si:ch211-215d8.2 | 0.298 | bzw1b | -0.056 |
| onecutl | 0.252 | sdc4 | -0.056 |
| onecut2 | 0.230 | aldob | -0.055 |
| lin28a | 0.226 | eef1da | -0.055 |
| onecut3b | 0.224 | nop58 | -0.053 |
| stmn3 | 0.222 | hspb1 | -0.052 |
| tubb2 | 0.218 | ucp2 | -0.052 |
| XLOC-013106 | 0.202 | dkc1 | -0.051 |
| cplx2l | 0.199 | ccng1 | -0.050 |
| si:dkey-10f21.4 | 0.195 | eif4ebp3l | -0.050 |
| snap25a | 0.195 | npm1a | -0.050 |
| map1b | 0.185 | banf1 | -0.049 |
| inab | 0.175 | fbl | -0.049 |
| stxbp1a | 0.169 | hdlbpa | -0.049 |
| nefmb | 0.161 | nop56 | -0.049 |
| stx1b | 0.161 | rsl1d1 | -0.049 |
| mllt11 | 0.158 | epcam | -0.048 |
| gap43 | 0.156 | msna | -0.048 |
| cplx2 | 0.154 | btg2 | -0.047 |
| elavl4 | 0.153 | lig1 | -0.046 |
| gng3 | 0.148 | si:ch211-286b5.5 | -0.046 |
| pcsk1nl | 0.148 | tuba8l | -0.045 |
| sncb | 0.147 | apoeb | -0.044 |
| stmn4l | 0.146 | ccnd1 | -0.044 |
| ywhag2 | 0.146 | cfl1l | -0.044 |
| rbfox3a | 0.145 | snu13b | -0.044 |
| stmn2b | 0.145 | stm | -0.044 |
| stmn2a | 0.144 | akap12b | -0.043 |
| id4 | 0.143 | cd63 | -0.043 |
| map1aa | 0.139 | cx43.4 | -0.043 |
| rbfox1 | 0.139 | zgc:162730 | -0.043 |
| syt2a | 0.139 | ahnak | -0.042 |
| tnr | 0.137 | chaf1a | -0.042 |