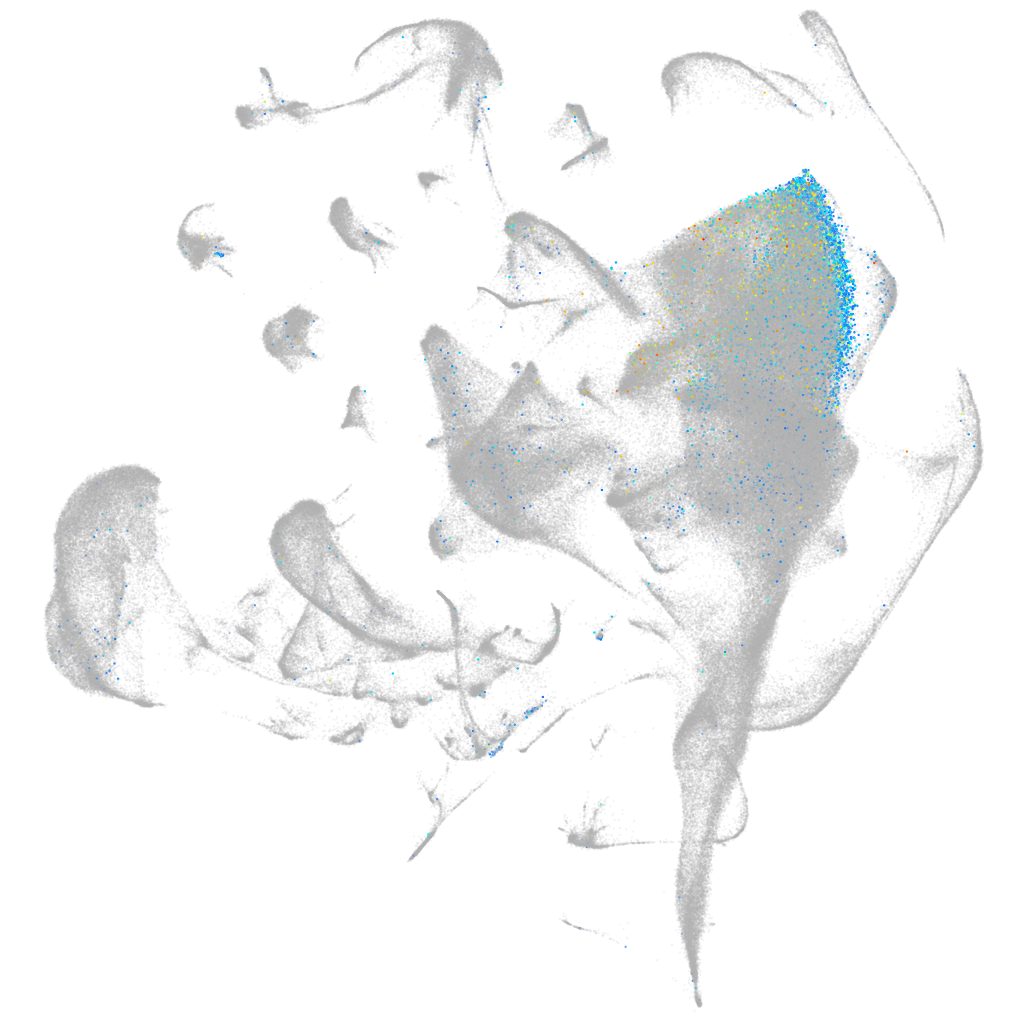si:ch211-215d8.2
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| onecut1 | 0.378 | id1 | -0.071 |
| onecut3a | 0.298 | sdc4 | -0.063 |
| XLOC-016165 | 0.270 | aldob | -0.062 |
| onecutl | 0.268 | eef1da | -0.061 |
| XLOC-013106 | 0.237 | vamp3 | -0.061 |
| tubb2 | 0.225 | nop58 | -0.057 |
| onecut2 | 0.216 | bzw1b | -0.056 |
| lin28a | 0.212 | ccng1 | -0.055 |
| onecut3b | 0.189 | dkc1 | -0.054 |
| snap25a | 0.181 | eif4ebp3l | -0.053 |
| map1b | 0.174 | hdlbpa | -0.053 |
| mllt11 | 0.173 | banf1 | -0.052 |
| cplx2l | 0.169 | ccnd1 | -0.052 |
| stxbp1a | 0.163 | fbl | -0.052 |
| gap43 | 0.162 | hspb1 | -0.052 |
| inab | 0.160 | npm1a | -0.052 |
| stmn3 | 0.158 | rsl1d1 | -0.052 |
| elavl3 | 0.157 | tuba8l | -0.052 |
| tmsb | 0.156 | nop56 | -0.051 |
| sncb | 0.155 | ucp2 | -0.051 |
| gng3 | 0.154 | epcam | -0.049 |
| tmeff1b | 0.154 | btg2 | -0.048 |
| elavl4 | 0.153 | snu13b | -0.048 |
| stmn1b | 0.149 | cfl1l | -0.047 |
| stmn2a | 0.149 | lig1 | -0.047 |
| erfl1 | 0.147 | msna | -0.047 |
| gng2 | 0.147 | nutf2l | -0.047 |
| stx1b | 0.146 | zgc:162730 | -0.047 |
| tubb5 | 0.146 | anp32b | -0.046 |
| vim | 0.146 | fam53b | -0.046 |
| cplx2 | 0.145 | fosab | -0.046 |
| map1aa | 0.145 | nop2 | -0.046 |
| syt2a | 0.145 | pcna | -0.046 |
| ywhag2 | 0.143 | ahnak | -0.045 |
| id4 | 0.142 | akap12b | -0.045 |