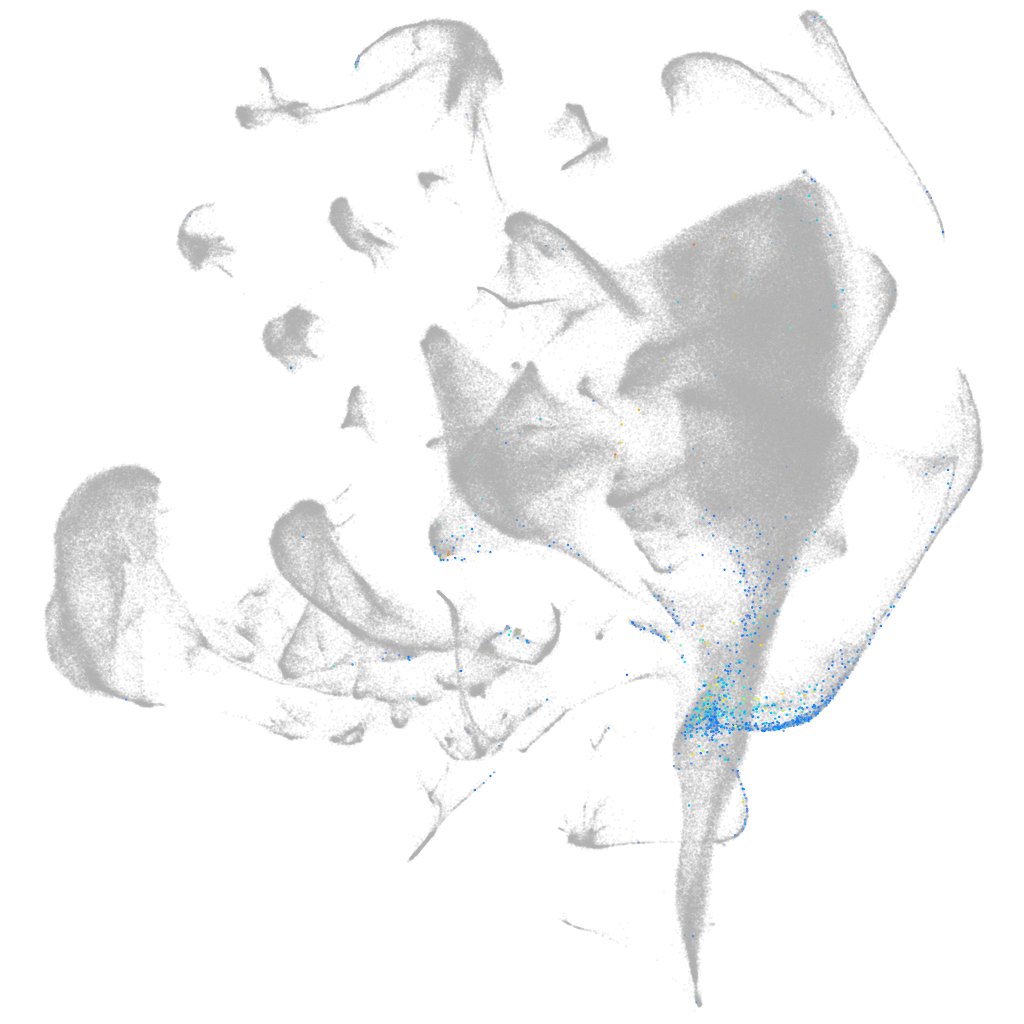
secreted frizzled-related protein 2 like
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| her1 | 0.185 | fabp3 | -0.045 |
| msgn1 | 0.176 | pvalb1 | -0.031 |
| BX001014.2 | 0.162 | pvalb2 | -0.030 |
| tbx16l | 0.162 | gpm6aa | -0.028 |
| cdx4 | 0.157 | celf2 | -0.027 |
| BX005254.3 | 0.155 | krt4 | -0.027 |
| her7 | 0.155 | tuba1c | -0.027 |
| hoxb7a | 0.146 | wu:fb55g09 | -0.027 |
| myf5 | 0.132 | ckbb | -0.026 |
| cdx1a | 0.124 | actc1b | -0.025 |
| hoxb10a | 0.119 | cyt1 | -0.025 |
| nradd | 0.110 | eif4a1b | -0.025 |
| hoxd10a | 0.105 | elavl3 | -0.025 |
| apoc1 | 0.103 | fabp7a | -0.025 |
| tbx6 | 0.102 | hbbe1.3 | -0.025 |
| vox | 0.102 | cotl1 | -0.024 |
| wnt5b | 0.099 | foxp4 | -0.024 |
| hoxa11a | 0.097 | gapdhs | -0.024 |
| hoxc3a | 0.097 | rtn1a | -0.024 |
| nid2a | 0.097 | cyt1l | -0.023 |
| ripply2 | 0.097 | eef1da | -0.023 |
| tbx16 | 0.095 | hbae3 | -0.023 |
| fgf8a | 0.093 | icn | -0.023 |
| hspb1 | 0.091 | mdkb | -0.023 |
| znfl1k | 0.091 | nova2 | -0.023 |
| BX005254.2 | 0.087 | sparc | -0.023 |
| tbxta | 0.087 | stmn1b | -0.023 |
| hoxc8a | 0.086 | tmsb | -0.023 |
| hoxd12a | 0.086 | marcksl1a | -0.023 |
| itm2cb | 0.084 | hbae1.1 | -0.022 |
| snai1a | 0.084 | mex3b | -0.022 |
| cx43.4 | 0.083 | mylpfa | -0.022 |
| hoxa11b | 0.083 | s100a10b | -0.022 |
| mllt3 | 0.083 | capns1a | -0.021 |
| vent | 0.083 | ccni | -0.021 |