secreted frizzled-related protein 2 like
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 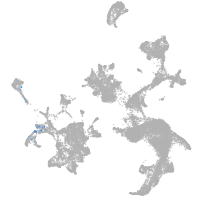 |
pronephros 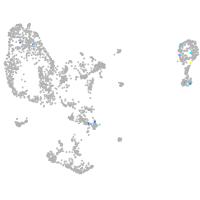 |
neural |
spinal cord / glia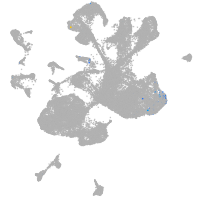 |
eye |
taste / olfactory |
otic / lateral line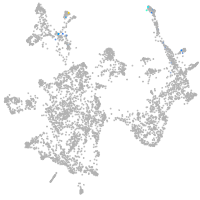 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hes6 | 0.223 | fabp3 | -0.135 |
| itm2cb | 0.219 | actc1b | -0.113 |
| hoxb7a | 0.209 | sparc | -0.111 |
| msgn1 | 0.197 | eef1da | -0.104 |
| cdx4 | 0.194 | rpl37 | -0.103 |
| her7 | 0.177 | ttn.2 | -0.099 |
| cx43.4 | 0.173 | bhmt | -0.097 |
| tbx16l | 0.172 | eif4a1b | -0.094 |
| vox | 0.170 | ak1 | -0.093 |
| wnt5b | 0.169 | zgc:114188 | -0.092 |
| BX005254.3 | 0.165 | pabpc4 | -0.092 |
| apoc1 | 0.163 | vdac3 | -0.092 |
| fzd10 | 0.163 | ckma | -0.091 |
| hoxc3a | 0.162 | ckmb | -0.090 |
| BX001014.2 | 0.158 | eno1a | -0.090 |
| tbx16 | 0.157 | tpi1b | -0.090 |
| nid2a | 0.155 | vwde | -0.090 |
| apoeb | 0.153 | atp2a1 | -0.090 |
| rarga | 0.145 | gamt | -0.090 |
| nradd | 0.145 | aldoab | -0.089 |
| rbm38 | 0.145 | tmem38a | -0.089 |
| cdx1a | 0.145 | rps10 | -0.089 |
| cyp26a1 | 0.141 | rps17 | -0.089 |
| her1 | 0.141 | ttn.1 | -0.089 |
| greb1 | 0.139 | mylpfa | -0.088 |
| vent | 0.138 | atp5if1b | -0.087 |
| tob1a | 0.137 | acta1b | -0.085 |
| her12 | 0.137 | tnnc2 | -0.085 |
| ube2e2 | 0.137 | gapdh | -0.083 |
| hoxa9a | 0.136 | col1a2 | -0.083 |
| si:ch73-281n10.2 | 0.134 | tpma | -0.082 |
| tagln3b | 0.133 | si:dkey-16p21.8 | -0.082 |
| ptgdsb.1 | 0.133 | klhl41b | -0.082 |
| dlc | 0.132 | gatm | -0.082 |
| hoxd9a | 0.131 | myl1 | -0.082 |




