homeobox A11a
ZFIN 
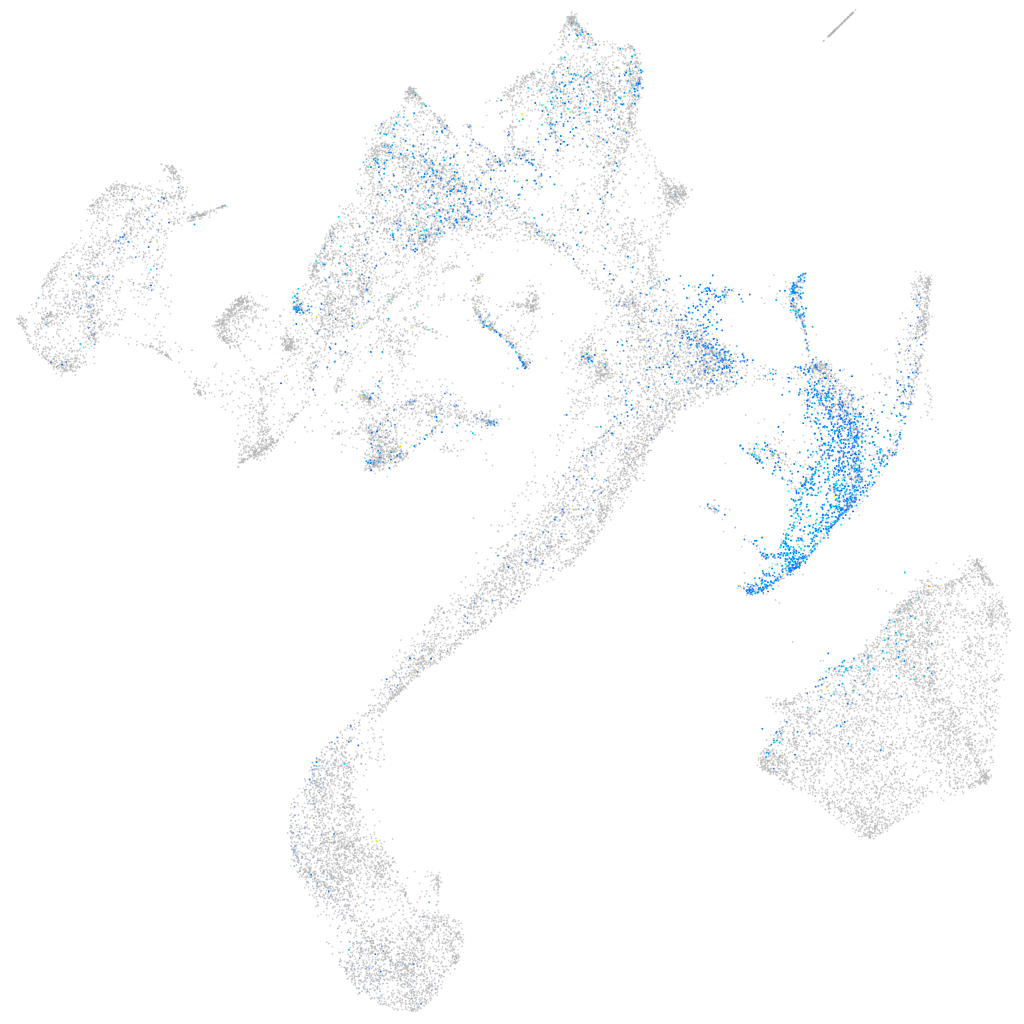
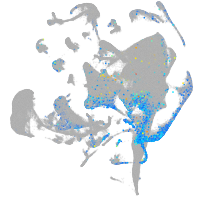
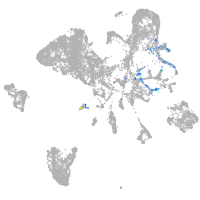
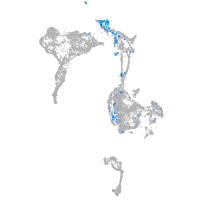
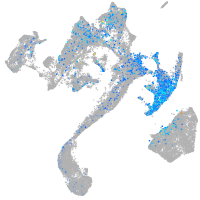
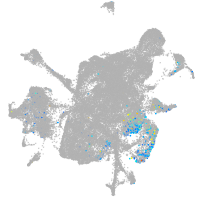
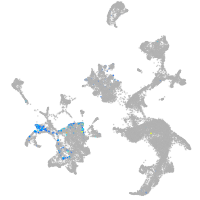
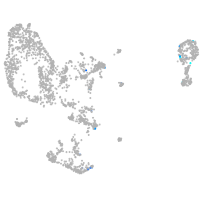
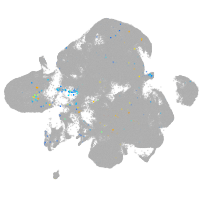
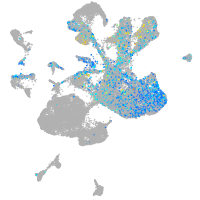
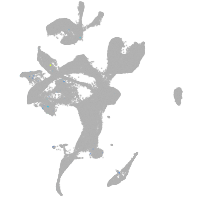
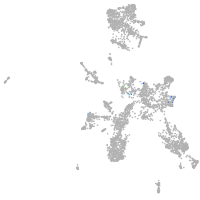
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hoxd10a | 0.451 | actc1b | -0.182 |
| hoxb10a | 0.421 | ttn.2 | -0.179 |
| hoxd12a | 0.419 | ak1 | -0.163 |
| hoxa11b | 0.394 | tmem38a | -0.162 |
| her1 | 0.384 | aldoab | -0.159 |
| hoxc11a | 0.383 | ttn.1 | -0.157 |
| itm2cb | 0.369 | atp2a1 | -0.157 |
| hoxa10b | 0.351 | pabpc4 | -0.156 |
| hoxd11a | 0.349 | gapdh | -0.156 |
| her12 | 0.341 | ckma | -0.156 |
| hoxc11b | 0.329 | ckmb | -0.154 |
| BX005254.3 | 0.323 | tnnc2 | -0.151 |
| her7 | 0.311 | klhl41b | -0.151 |
| pcdh8 | 0.302 | desma | -0.150 |
| sall4 | 0.297 | srl | -0.149 |
| fbln1 | 0.294 | eno1a | -0.148 |
| hoxc10a | 0.290 | acta1b | -0.147 |
| hoxd3a | 0.288 | ldb3a | -0.146 |
| rarga | 0.287 | mybphb | -0.146 |
| ephb3 | 0.286 | mylpfa | -0.144 |
| hoxd9a | 0.285 | neb | -0.144 |
| hes6 | 0.278 | tpma | -0.142 |
| greb1 | 0.273 | myl1 | -0.142 |
| nid2a | 0.268 | fxr2 | -0.141 |
| msgn1 | 0.262 | cav3 | -0.141 |
| LO016987.2 | 0.254 | txlnbb | -0.140 |
| apoc1 | 0.251 | acta1a | -0.139 |
| XLOC-042222 | 0.251 | gamt | -0.139 |
| tbx16l | 0.249 | nme2b.2 | -0.138 |
| gdf11 | 0.246 | CABZ01078594.1 | -0.138 |
| zgc:162939 | 0.244 | zgc:101853 | -0.137 |
| hnrnpa0l | 0.242 | gatm | -0.137 |
| hoxa13b | 0.241 | cox17 | -0.136 |
| tspan7 | 0.239 | FQ323156.1 | -0.135 |
| hoxb7a | 0.238 | tnni2b.1 | -0.135 |