homeobox A11a
ZFIN 
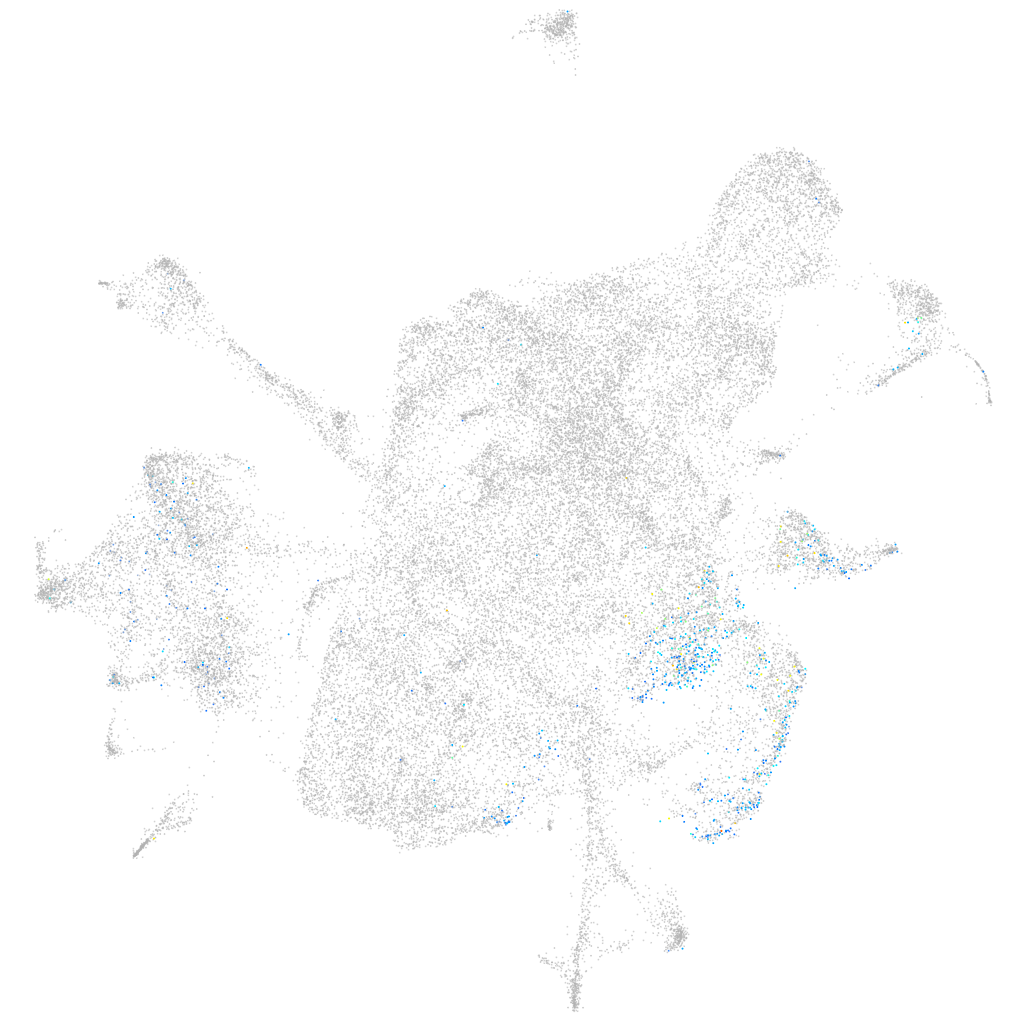
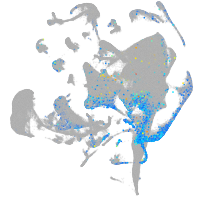
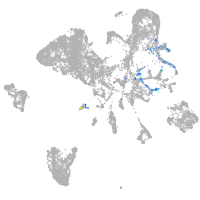
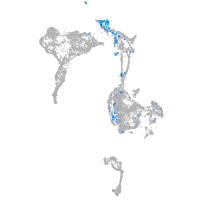
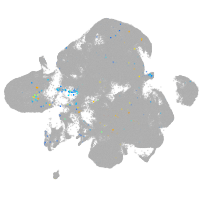
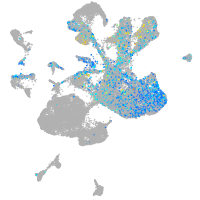
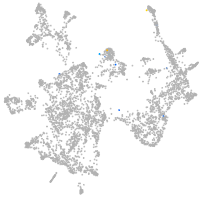
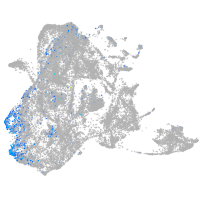
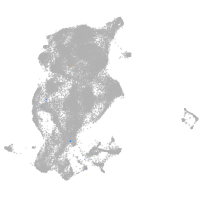
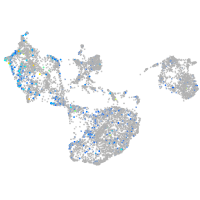
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hoxc11a | 0.297 | foxc1b | -0.054 |
| hoxc11b | 0.216 | col9a1a | -0.054 |
| hoxc10a | 0.214 | col2a1a | -0.048 |
| hoxa11b | 0.207 | col9a2 | -0.046 |
| hoxd11a | 0.188 | rarga | -0.045 |
| hoxb9a | 0.185 | dlx5a | -0.043 |
| hoxa10b | 0.184 | gsc | -0.042 |
| hoxd10a | 0.181 | six1a | -0.042 |
| hoxd12a | 0.173 | barx1 | -0.041 |
| emilin2b | 0.172 | ecrg4a | -0.041 |
| hoxd9a | 0.168 | col9a3 | -0.041 |
| hoxb10a | 0.164 | cldn11a | -0.040 |
| hoxa9a | 0.154 | six2a | -0.040 |
| hoxc12b | 0.144 | adgrl2a | -0.039 |
| aplnr2 | 0.140 | col11a2 | -0.039 |
| evx1 | 0.135 | cnmd | -0.039 |
| hoxa9b | 0.135 | prdx2 | -0.037 |
| zmp:0000000846 | 0.130 | fxyd1 | -0.036 |
| BX005254.3 | 0.130 | dlx2a | -0.035 |
| postnb | 0.129 | sox9a | -0.035 |
| kazald2 | 0.128 | hpgd | -0.035 |
| apela | 0.126 | LO018188.1 | -0.035 |
| c1qtnf12 | 0.125 | nfia | -0.035 |
| hoxc6b | 0.119 | CABZ01092746.1 | -0.035 |
| tuba8l2 | 0.117 | fgfbp2b | -0.034 |
| col5a2a | 0.113 | ucmab | -0.034 |
| hoxa13a | 0.106 | junba | -0.034 |
| cfd | 0.105 | VIT | -0.034 |
| tnn | 0.104 | lsm6 | -0.034 |
| clec19a | 0.103 | KCTD15 | -0.033 |
| fstl1a | 0.102 | sema3bl | -0.033 |
| cd82a | 0.099 | fibina | -0.033 |
| hoxc9a | 0.099 | dlx4a | -0.032 |
| fbln1 | 0.098 | atp5mc3a | -0.032 |
| arl4ca | 0.097 | sfrp2 | -0.032 |