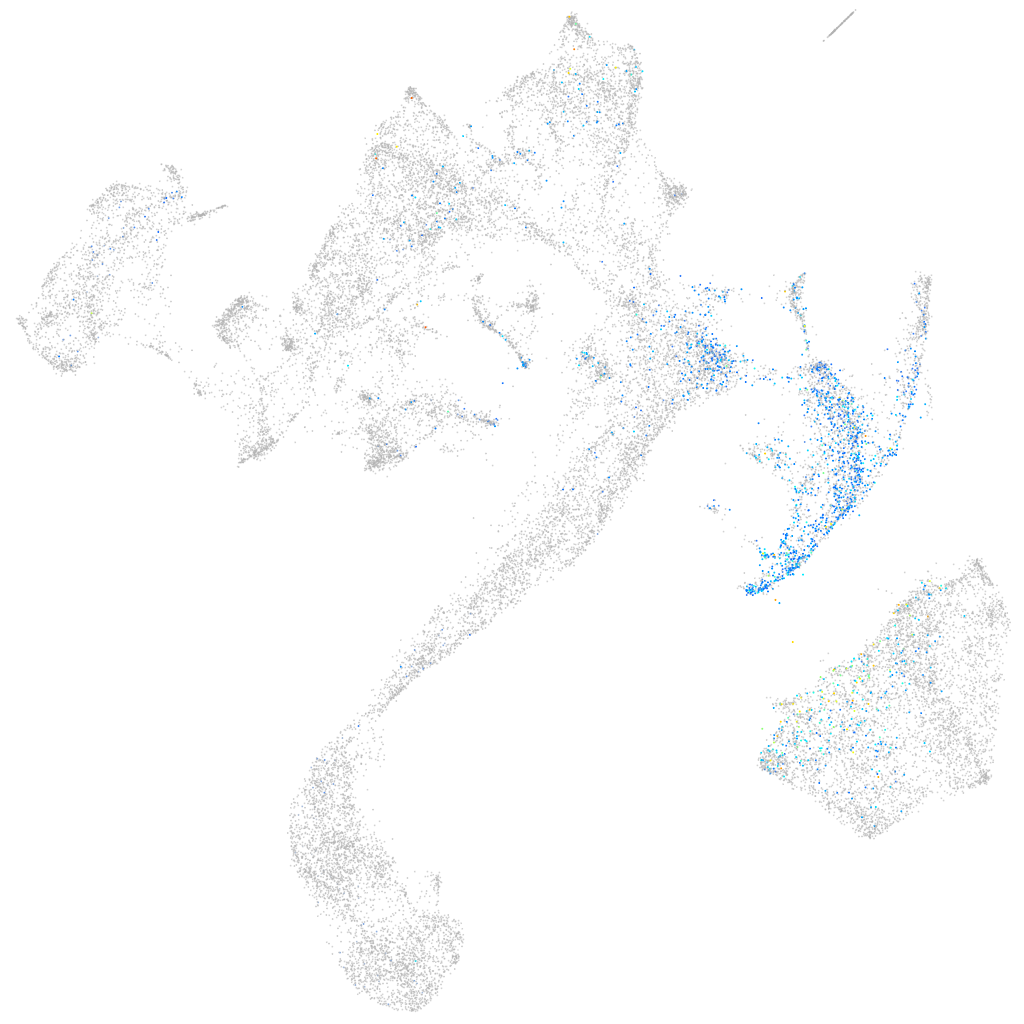
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 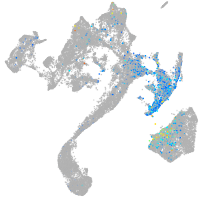 |
mural cells / non-skeletal muscle 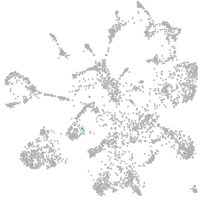 |
mesenchyme 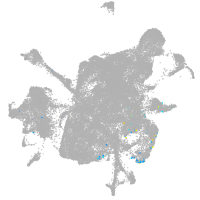 |
hematopoietic / vasculature 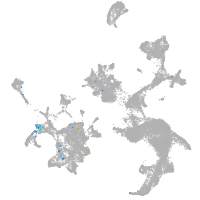 |
pronephros 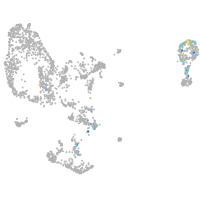 |
neural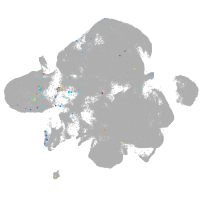 |
spinal cord / glia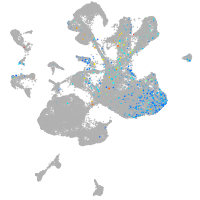 |
eye |
taste / olfactory |
otic / lateral line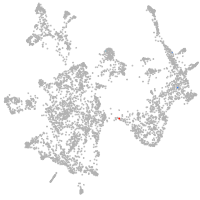 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hoxb10a | 0.300 | fabp3 | -0.174 |
| BX005254.3 | 0.281 | actc1b | -0.159 |
| hoxd10a | 0.278 | ttn.2 | -0.138 |
| hes6 | 0.273 | pabpc4 | -0.138 |
| itm2cb | 0.269 | ak1 | -0.136 |
| hoxb7a | 0.256 | sparc | -0.135 |
| her12 | 0.249 | atp2a1 | -0.135 |
| her1 | 0.244 | eno1a | -0.135 |
| hoxa10b | 0.231 | ckma | -0.133 |
| her7 | 0.225 | ckmb | -0.133 |
| BX001014.2 | 0.223 | tmem38a | -0.133 |
| hoxa11b | 0.222 | bhmt | -0.130 |
| apoc1 | 0.221 | eef1da | -0.129 |
| cx43.4 | 0.219 | aldoab | -0.128 |
| pcdh8 | 0.217 | mylpfa | -0.128 |
| cdx4 | 0.216 | gamt | -0.126 |
| sall4 | 0.213 | gapdh | -0.126 |
| hoxc3a | 0.208 | tnnc2 | -0.123 |
| fgf8a | 0.208 | acta1b | -0.123 |
| hoxd12a | 0.207 | srl | -0.120 |
| nid2a | 0.205 | myl1 | -0.120 |
| hoxd9a | 0.205 | tpi1b | -0.118 |
| hoxa11a | 0.202 | klhl41b | -0.117 |
| apoeb | 0.202 | neb | -0.117 |
| greb1 | 0.200 | FQ323156.1 | -0.117 |
| wnt5b | 0.197 | desma | -0.116 |
| hoxd11a | 0.196 | pvalb1 | -0.115 |
| cyp26a1 | 0.193 | tpma | -0.115 |
| ptgdsb.1 | 0.187 | ldb3a | -0.115 |
| gdf11 | 0.187 | cav3 | -0.115 |
| hoxa9a | 0.185 | ldb3b | -0.114 |
| hoxa9b | 0.184 | gatm | -0.114 |
| vox | 0.183 | mylz3 | -0.114 |
| si:ch73-281n10.2 | 0.181 | eif4a1b | -0.114 |
| selenon | 0.179 | pvalb2 | -0.114 |