"wingless-type MMTV integration site family, member 2Bb"
ZFIN 
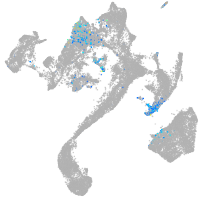
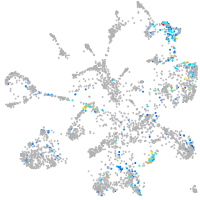
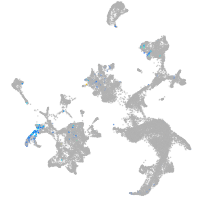
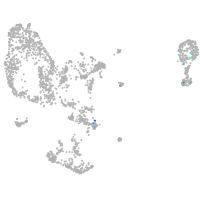
Other cell groups


Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| fgf10b | 0.507 | aldob | -0.095 |
| isl2a | 0.491 | eef1da | -0.091 |
| hoxb10a | 0.447 | gapdh | -0.090 |
| pax2a | 0.398 | ahcy | -0.089 |
| ptgs1 | 0.390 | eno3 | -0.089 |
| hoxa11b | 0.380 | sod1 | -0.088 |
| hapln1b | 0.370 | gamt | -0.080 |
| BX005254.3 | 0.369 | atp5if1b | -0.073 |
| hoxc8a | 0.355 | tpi1b | -0.072 |
| sall4 | 0.333 | gatm | -0.071 |
| fbln1 | 0.321 | fabp3 | -0.070 |
| hoxd9a | 0.316 | gstp1 | -0.070 |
| hoxa10b | 0.315 | fbp1b | -0.069 |
| nkx1.2la | 0.315 | apoa1b | -0.068 |
| rspo2 | 0.314 | mdh1aa | -0.067 |
| hoxb9a | 0.312 | glud1b | -0.066 |
| hoxc9a | 0.311 | gstt1a | -0.066 |
| msx3 | 0.309 | aldh7a1 | -0.066 |
| wdr66 | 0.302 | apoa4b.1 | -0.065 |
| apela | 0.300 | bhmt | -0.065 |
| hoxc11a | 0.298 | nme2b.1 | -0.065 |
| hoxd10a | 0.297 | prdx2 | -0.065 |
| hoxc3a | 0.287 | pgk1 | -0.065 |
| hoxb7a | 0.282 | BX908782.3 | -0.065 |
| evx1 | 0.282 | suclg1 | -0.064 |
| hoxa9b | 0.276 | nupr1b | -0.064 |
| hoxc6b | 0.274 | ppa1b | -0.064 |
| hoxd11a | 0.274 | apoc2 | -0.064 |
| BX005254.2 | 0.270 | lgals2b | -0.064 |
| sp7 | 0.258 | sod2 | -0.064 |
| fgfrl1b | 0.257 | gpx4a | -0.064 |
| zgc:171775 | 0.256 | cox6b1 | -0.063 |
| kif26ba | 0.255 | pklr | -0.062 |
| ndnf | 0.250 | prdx6 | -0.062 |
| gdnfa | 0.250 | mat1a | -0.062 |