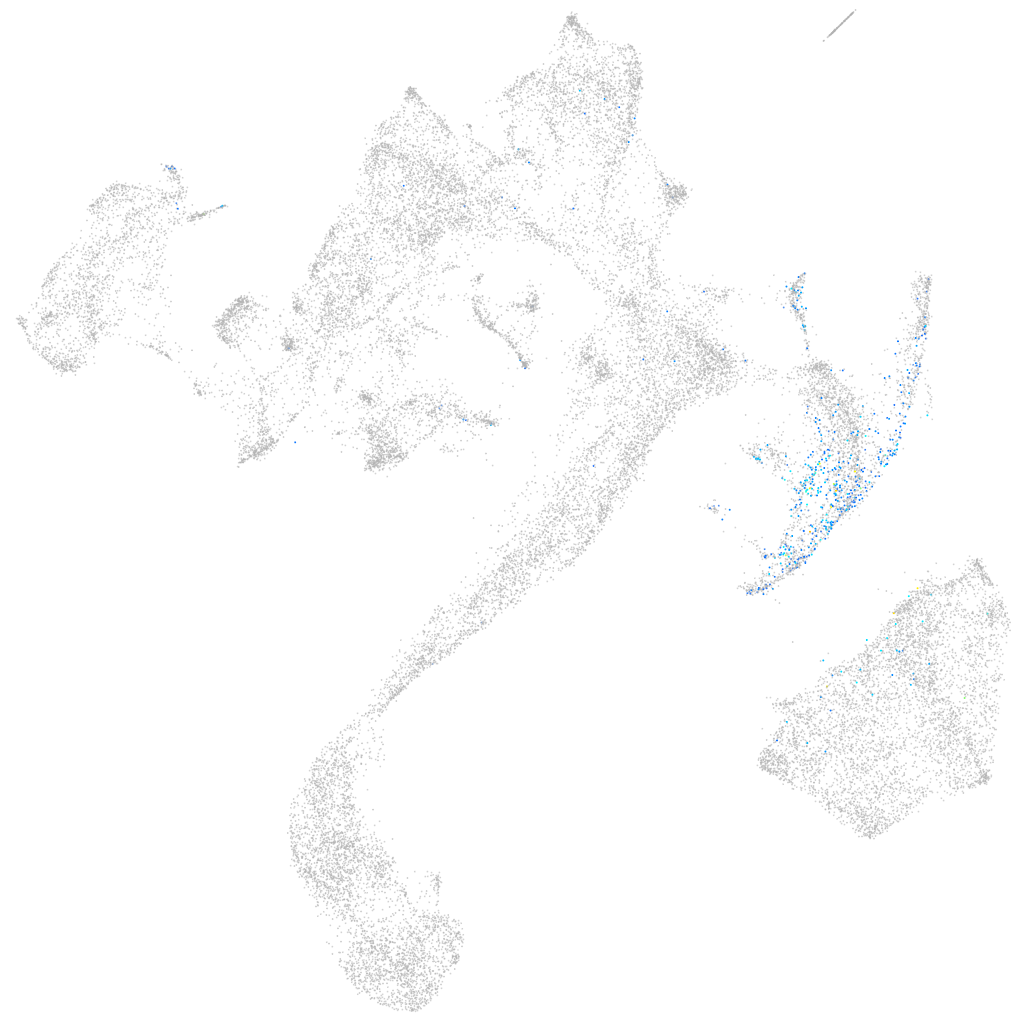
"5-hydroxytryptamine (serotonin) receptor 5A, genome duplicate a"
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| itm2cb | 0.278 | fabp3 | -0.108 |
| hoxd10a | 0.255 | sparc | -0.089 |
| her1 | 0.250 | actc1b | -0.082 |
| her7 | 0.247 | eef1da | -0.081 |
| hoxb10a | 0.242 | pabpc4 | -0.079 |
| her12 | 0.236 | bhmt | -0.077 |
| msgn1 | 0.224 | vwde | -0.075 |
| thbs2a | 0.223 | ckma | -0.075 |
| tbx16l | 0.220 | FQ323156.1 | -0.074 |
| pcdh8 | 0.218 | atp2a1 | -0.074 |
| tnfrsf9a | 0.214 | ckmb | -0.074 |
| hes6 | 0.212 | ttn.2 | -0.072 |
| hoxd12a | 0.199 | ak1 | -0.072 |
| nid2a | 0.191 | rplp2 | -0.071 |
| BX005254.3 | 0.180 | gapdh | -0.070 |
| ephb3 | 0.179 | mylpfa | -0.070 |
| fzd10 | 0.177 | gamt | -0.070 |
| hoxa11a | 0.175 | tmem38a | -0.069 |
| hoxd11a | 0.175 | si:dkey-16p21.8 | -0.068 |
| tob1a | 0.170 | COX3 | -0.068 |
| hoxc11a | 0.169 | cox17 | -0.068 |
| sall4 | 0.169 | eno1a | -0.067 |
| snai1a | 0.167 | nme2b.2 | -0.067 |
| rarga | 0.165 | pvalb1 | -0.066 |
| alpi.1 | 0.165 | myl1 | -0.066 |
| cyp26a1 | 0.161 | pvalb2 | -0.065 |
| gdf11 | 0.161 | aldoab | -0.065 |
| phkg2 | 0.161 | col1a2 | -0.065 |
| hoxb7a | 0.160 | mylz3 | -0.064 |
| hoxa11b | 0.160 | srl | -0.063 |
| hoxa10b | 0.160 | neb | -0.063 |
| zgc:162939 | 0.159 | gatm | -0.063 |
| greb1 | 0.158 | tnnt3a | -0.063 |
| LO016987.2 | 0.158 | ldb3b | -0.063 |
| fn1a | 0.156 | tnnc2 | -0.062 |