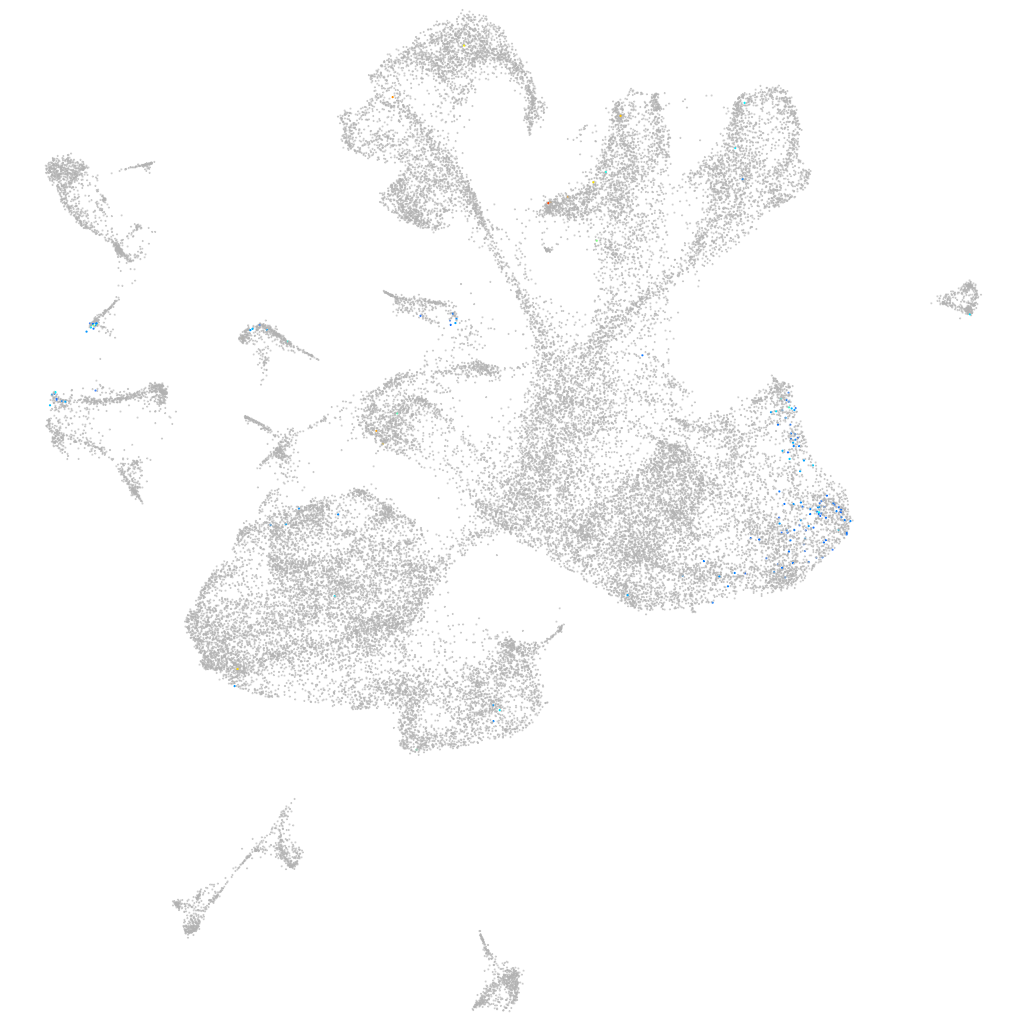
"5-hydroxytryptamine (serotonin) receptor 5A, genome duplicate a"
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| apela | 0.122 | fabp3 | -0.060 |
| BX548073.1 | 0.120 | tmsb4x | -0.040 |
| sp5l | 0.119 | marcksl1b | -0.040 |
| si:ch73-236c18.3 | 0.115 | fabp7a | -0.035 |
| cyp21a2 | 0.112 | gpm6aa | -0.034 |
| igl4v9 | 0.112 | CU467822.1 | -0.033 |
| FP016005.1 | 0.110 | si:ch73-21g5.7 | -0.032 |
| pou5f3 | 0.104 | ncam1a | -0.032 |
| cdx4 | 0.102 | vim | -0.030 |
| LO017682.1 | 0.102 | nova2 | -0.030 |
| RF00325 | 0.101 | tmsb | -0.029 |
| LOC100536326 | 0.100 | si:ch211-57n23.4 | -0.028 |
| ntd5 | 0.099 | rtn1a | -0.028 |
| cxcr4a | 0.098 | pou3f1 | -0.028 |
| CT033825.1 | 0.098 | zgc:165461 | -0.027 |
| rp1l1b | 0.098 | elavl3 | -0.027 |
| lamb1a | 0.094 | pou3f3b | -0.027 |
| XLOC-042222 | 0.092 | ckbb | -0.027 |
| chrd | 0.091 | tspan3a | -0.026 |
| fgf8a | 0.090 | fgfr3 | -0.026 |
| si:dkey-165n16.5 | 0.088 | nr2f1a | -0.026 |
| nkx1.2la | 0.088 | nat8l | -0.025 |
| nradd | 0.088 | CU634008.1 | -0.025 |
| LOC110439867 | 0.087 | plp1a | -0.025 |
| hspb1 | 0.086 | gpm6bb | -0.025 |
| prdm1c | 0.085 | LOC798783 | -0.025 |
| hoxd10a | 0.084 | hmgb3a | -0.025 |
| apoc1 | 0.082 | myt1a | -0.025 |
| serpinh1b | 0.082 | rnasekb | -0.025 |
| col15a1a | 0.079 | si:ch211-133n4.4 | -0.024 |
| marveld2b | 0.079 | lrrn1 | -0.024 |
| BX001014.2 | 0.079 | meis1b | -0.024 |
| SPATA1 | 0.078 | cd82a | -0.024 |
| si:dkey-29m1.2 | 0.077 | efcc1 | -0.024 |
| LOC108179174 | 0.077 | slc6a9 | -0.024 |