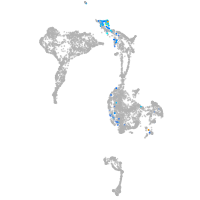
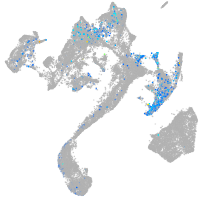
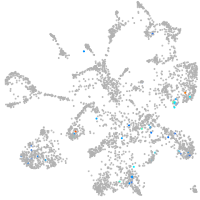
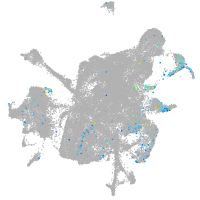
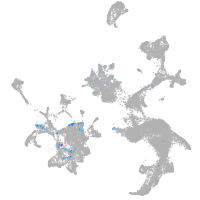
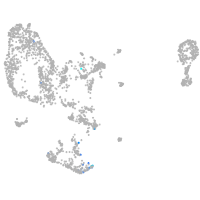
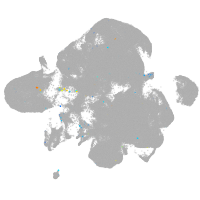
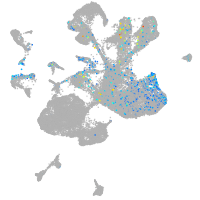
homeobox D13a
ZFIN 
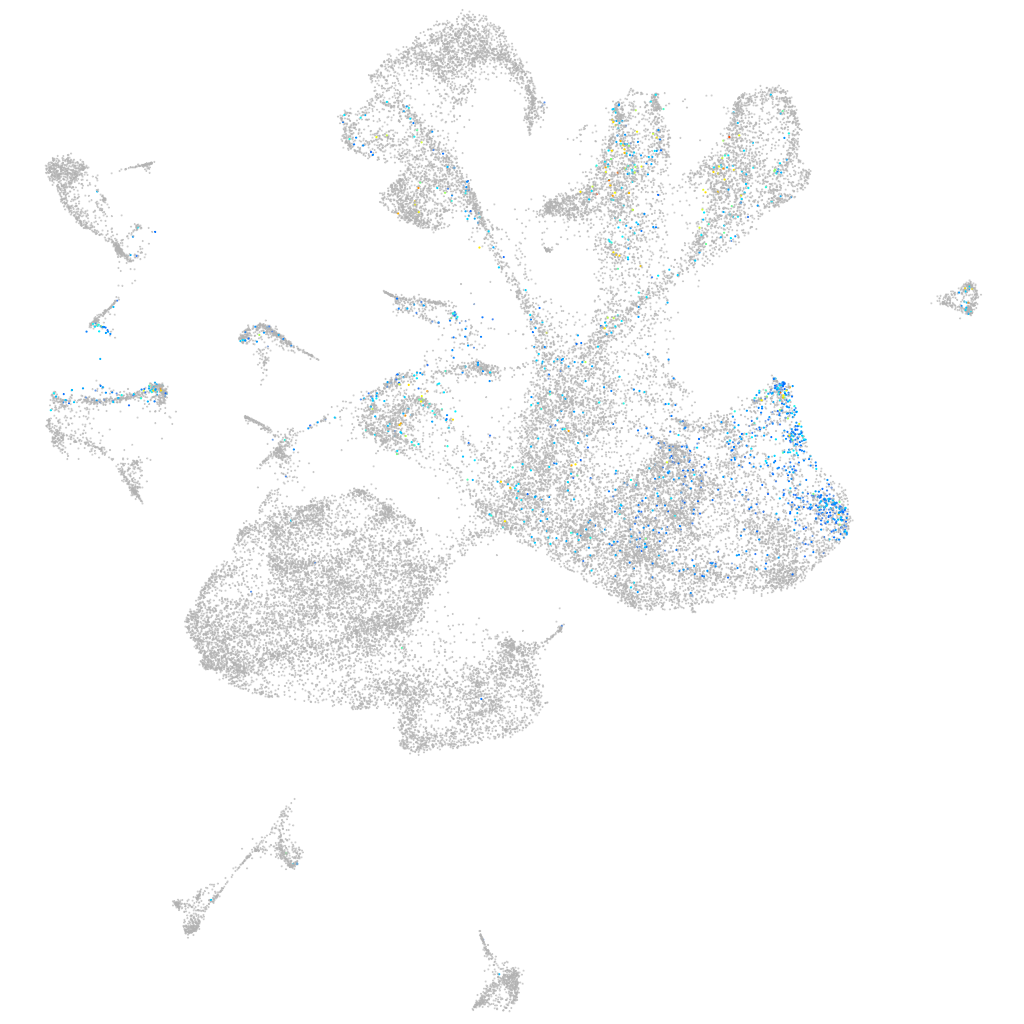
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hoxd12a | 0.335 | tmsb4x | -0.079 |
| hoxc13a | 0.278 | fabp7a | -0.079 |
| hoxc13b | 0.240 | fabp3 | -0.066 |
| hoxc12b | 0.233 | zgc:165461 | -0.059 |
| hoxa13b | 0.221 | CR848047.1 | -0.058 |
| hoxa13a | 0.202 | gpm6bb | -0.056 |
| hoxb13a | 0.191 | CU467822.1 | -0.055 |
| hoxc11a | 0.188 | slc1a2b | -0.054 |
| eve1 | 0.180 | marcksl1a | -0.053 |
| hoxc11b | 0.176 | marcksl1b | -0.051 |
| pou5f3 | 0.171 | ppap2d | -0.051 |
| hoxa11a | 0.171 | CU634008.1 | -0.049 |
| sp5l | 0.167 | slc6a1b | -0.047 |
| hoxd10a | 0.166 | si:dkey-85k7.7 | -0.047 |
| hoxc12a | 0.164 | atp1a1b | -0.047 |
| si:dkeyp-110a12.4 | 0.162 | hoxc1a | -0.046 |
| hoxa10b | 0.154 | klf6a | -0.046 |
| smkr1 | 0.151 | CR751602.2 | -0.045 |
| dnase1l4.1 | 0.148 | efhd1 | -0.045 |
| hoxa11b | 0.146 | si:ch73-21g5.7 | -0.044 |
| apoc1 | 0.145 | dhrs3b | -0.044 |
| hoxd11a | 0.144 | phox2bb | -0.043 |
| SPATA1 | 0.137 | glula | -0.043 |
| tppp3 | 0.135 | crabp1a | -0.042 |
| noto | 0.135 | fgfr2 | -0.042 |
| hoxb10a | 0.135 | mdkb | -0.042 |
| nkx1.2la | 0.132 | plp1a | -0.042 |
| si:dkey-18j18.3 | 0.126 | nfia | -0.042 |
| c8h22orf15 | 0.124 | ca4a | -0.041 |
| daw1 | 0.123 | ckbb | -0.041 |
| cdx4 | 0.123 | celf2 | -0.041 |
| krt18a.1 | 0.122 | meis2b | -0.041 |
| mllt3 | 0.122 | cyp26b1 | -0.040 |
| foxd5 | 0.122 | cxcl12a | -0.040 |
| spag6 | 0.121 | atp1b4 | -0.039 |