homeobox C11b
ZFIN 
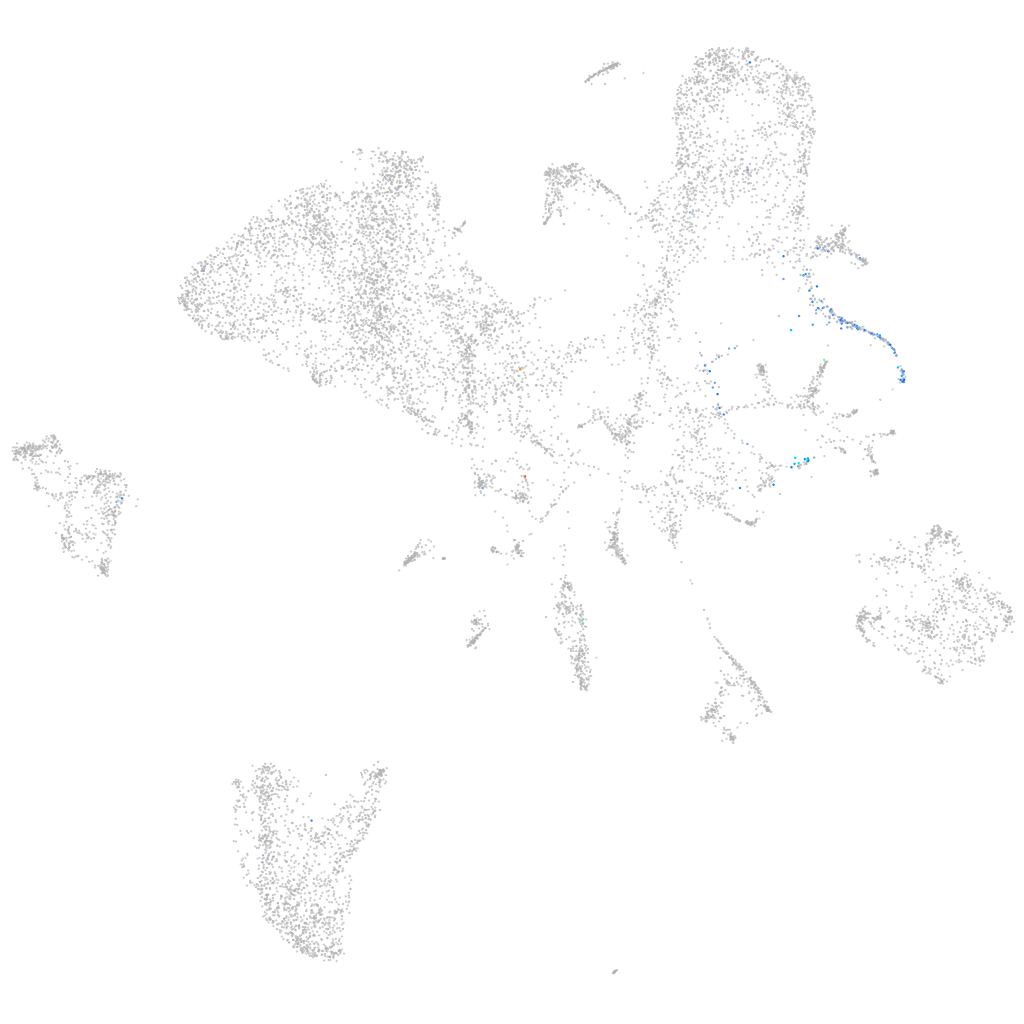
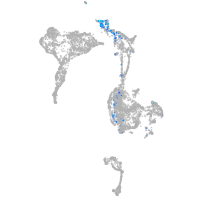
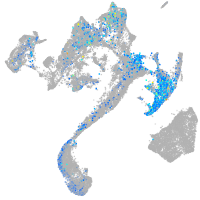
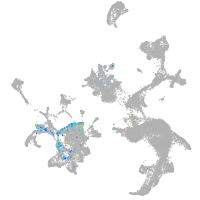
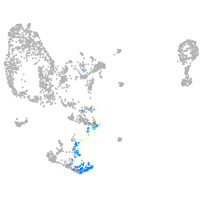
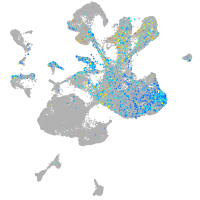
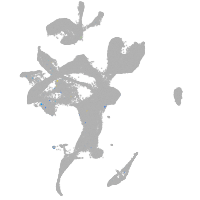
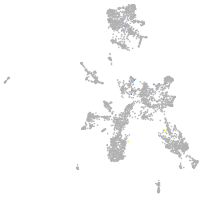
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| evx1 | 0.373 | gatm | -0.077 |
| si:dkey-245n4.2 | 0.318 | gamt | -0.075 |
| cdh16 | 0.308 | bhmt | -0.075 |
| hoxa13a | 0.305 | mat1a | -0.071 |
| hoxb13a | 0.262 | aqp12 | -0.068 |
| hoxd13a | 0.256 | apoc2 | -0.065 |
| hoxc11a | 0.249 | nupr1b | -0.063 |
| hoxd12a | 0.228 | cdo1 | -0.062 |
| hoxb10a | 0.224 | agxtb | -0.062 |
| emx1 | 0.213 | gcshb | -0.062 |
| sp8a | 0.211 | kng1 | -0.061 |
| etf1a | 0.211 | grhprb | -0.060 |
| trim35-12 | 0.211 | afp4 | -0.060 |
| hoxc13a | 0.209 | apoa1b | -0.060 |
| sstr5 | 0.209 | pnp4b | -0.059 |
| hoxc10a | 0.204 | scp2a | -0.059 |
| isl2a | 0.197 | si:dkey-16p21.8 | -0.059 |
| ponzr1 | 0.195 | ahcy | -0.058 |
| zgc:64022 | 0.194 | dap | -0.058 |
| malb | 0.186 | cx32.3 | -0.057 |
| si:ch211-153b23.4 | 0.185 | serpinf2b | -0.057 |
| LOC103909163 | 0.185 | slco1d1 | -0.057 |
| ftr83 | 0.184 | abat | -0.057 |
| CABZ01069184.1 | 0.182 | apoa4b.1 | -0.056 |
| stk25a | 0.182 | rbp2b | -0.056 |
| CABZ01025162.1 | 0.181 | cpn1 | -0.055 |
| noxo1b | 0.181 | ttc36 | -0.055 |
| areg | 0.179 | rltgr | -0.055 |
| efcc1 | 0.176 | suclg2 | -0.054 |
| hoxa13b | 0.176 | etnppl | -0.054 |
| ca4c | 0.173 | mgst1.2 | -0.054 |
| hoxa11a | 0.170 | hpda | -0.054 |
| si:dkey-92i15.4 | 0.169 | msrb2 | -0.054 |
| FP103048.1 | 0.169 | gapdh | -0.054 |
| CU929379.1 | 0.167 | serpina1l | -0.053 |