eukaryotic translation termination factor 1a
ZFIN 
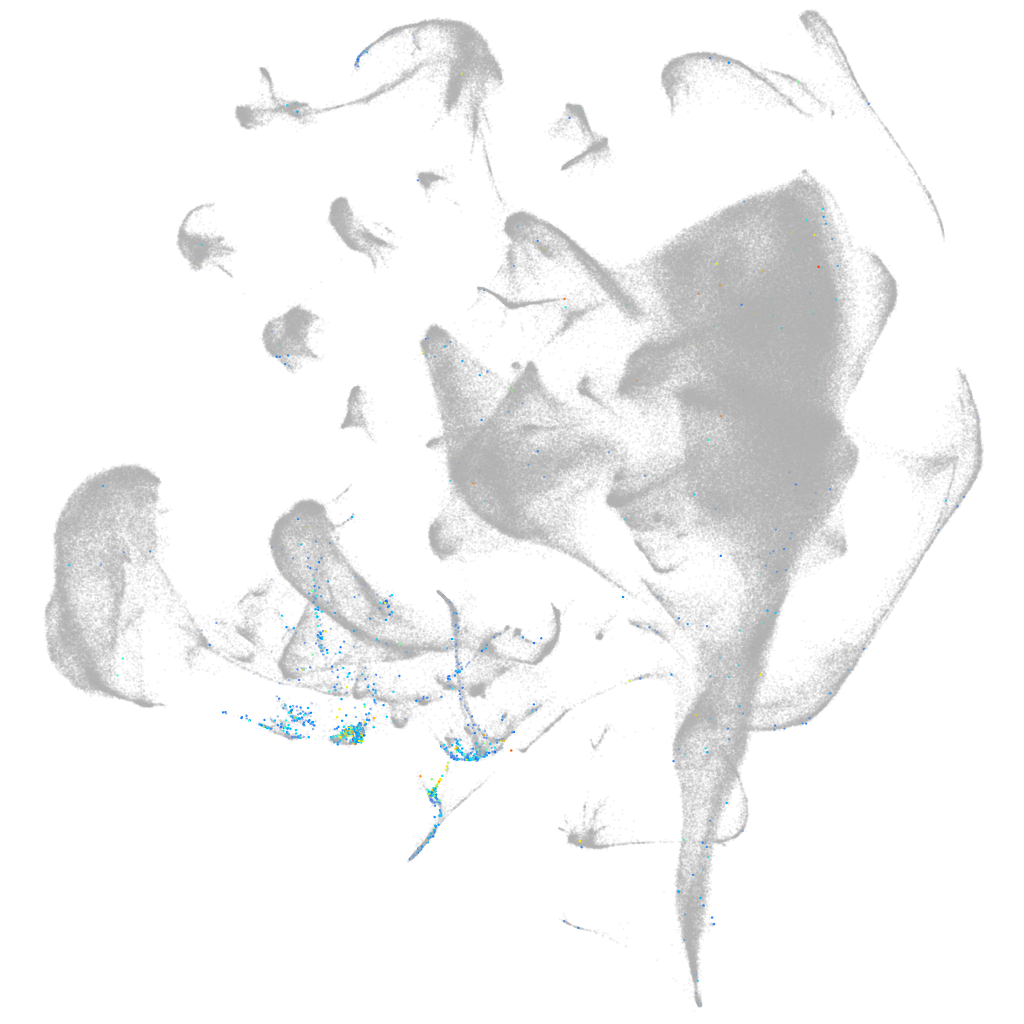
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm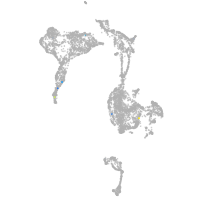 |
muscle  |
mural cells / non-skeletal muscle 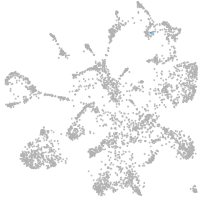 |
mesenchyme  |
hematopoietic / vasculature 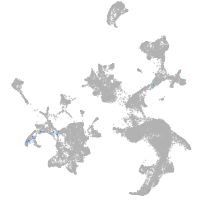 |
pronephros 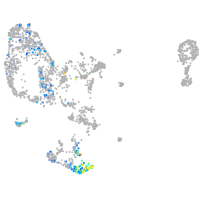 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:dkey-192d15.2 | 0.412 | marcksl1a | -0.051 |
| foxi3a | 0.406 | gpm6aa | -0.046 |
| sstr5 | 0.390 | tuba1c | -0.045 |
| gcm2 | 0.372 | nova2 | -0.044 |
| ca15a | 0.343 | rtn1a | -0.043 |
| mal2 | 0.340 | gpm6ab | -0.042 |
| si:dkey-33i11.4 | 0.311 | atp6v0cb | -0.040 |
| foxi3b | 0.300 | hmgb1b | -0.039 |
| ceacam1 | 0.288 | celf2 | -0.038 |
| rhcgb | 0.279 | ckbb | -0.038 |
| si:ch211-270g19.5 | 0.260 | tubb5 | -0.038 |
| agrp | 0.256 | tuba1a | -0.036 |
| sgk2a | 0.252 | elavl3 | -0.035 |
| trim35-12 | 0.243 | mdkb | -0.035 |
| si:dkeyp-97e7.9 | 0.232 | si:ch211-133n4.4 | -0.035 |
| cldnh | 0.220 | cadm3 | -0.034 |
| si:ch73-14h1.2 | 0.220 | cspg5a | -0.034 |
| slc4a1b | 0.219 | fam168a | -0.033 |
| zgc:193726 | 0.215 | stmn1b | -0.033 |
| zgc:56622 | 0.215 | actc1b | -0.032 |
| atp1b1b | 0.210 | tmeff1b | -0.032 |
| CR356236.1 | 0.207 | atpv0e2 | -0.031 |
| KCNJ15 | 0.189 | cd81a | -0.031 |
| prkag3b | 0.187 | fabp7a | -0.031 |
| prg4a | 0.185 | mdka | -0.031 |
| sat1a.1 | 0.175 | cdh2 | -0.030 |
| cox5b2 | 0.174 | rtn1b | -0.030 |
| CU571336.1 | 0.173 | si:ch211-137a8.4 | -0.030 |
| si:ch73-359m17.9 | 0.172 | atp1a3a | -0.029 |
| atp6v0a1a | 0.168 | atp1b2a | -0.029 |
| tmprss2 | 0.168 | cd99l2 | -0.029 |
| ca2 | 0.167 | hmgb3a | -0.029 |
| si:dkey-251i10.1 | 0.167 | hnrnpa0a | -0.029 |
| rasgrp4 | 0.163 | ncam1a | -0.029 |
| prom2 | 0.162 | sncb | -0.029 |