eukaryotic translation termination factor 1a
ZFIN 
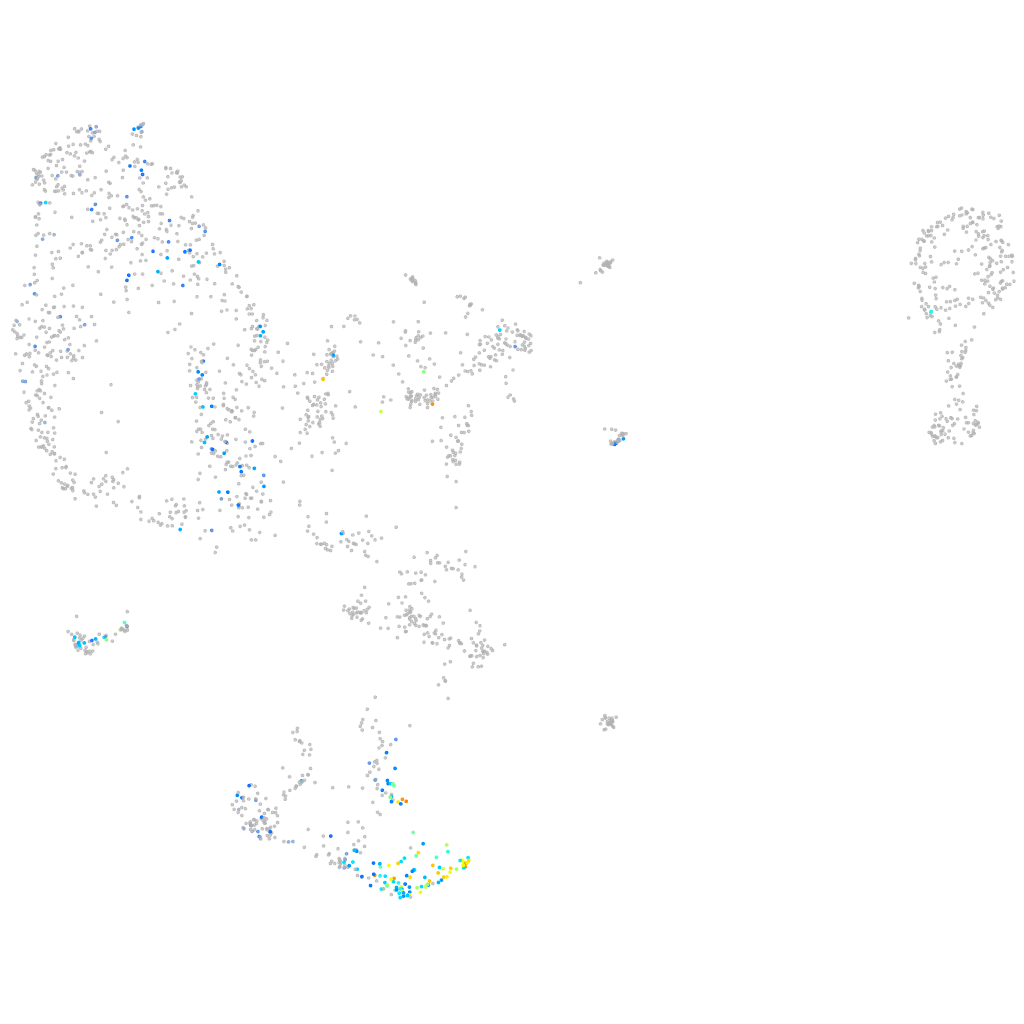
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm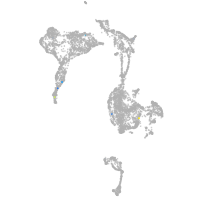 |
muscle  |
mural cells / non-skeletal muscle 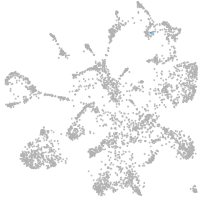 |
mesenchyme  |
hematopoietic / vasculature 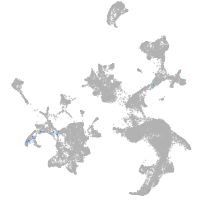 |
pronephros 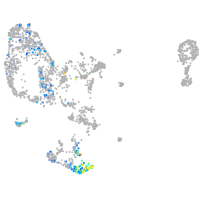 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| atp1a1a.5 | 0.607 | acy3.2 | -0.167 |
| si:dkey-36i7.3 | 0.595 | eif5a2 | -0.156 |
| slc12a10.3 | 0.592 | hspb1 | -0.153 |
| slc9a3.2 | 0.588 | marcksl1b | -0.152 |
| atp1a1a.3 | 0.581 | si:ch73-1a9.3 | -0.151 |
| trim35-12 | 0.546 | marcksb | -0.150 |
| si:ch211-153b23.5 | 0.534 | snrpg | -0.146 |
| soul2 | 0.529 | msna | -0.145 |
| fabp2 | 0.527 | si:dkey-151g10.6 | -0.143 |
| degs2 | 0.525 | si:dkey-194e6.1 | -0.139 |
| si:ch211-79k12.1 | 0.522 | apoeb | -0.138 |
| rps6ka5 | 0.517 | pdzk1 | -0.137 |
| ca4c | 0.509 | wu:fb97g03 | -0.136 |
| mal2 | 0.508 | hnrnpabb | -0.134 |
| aspg | 0.506 | grhprb | -0.133 |
| tspan13a | 0.496 | snrpf | -0.133 |
| agr2 | 0.484 | ptmab | -0.132 |
| cldne | 0.476 | ftcd | -0.132 |
| rnf183 | 0.472 | cx43.4 | -0.131 |
| zgc:158423 | 0.470 | rps12 | -0.130 |
| zgc:92275 | 0.468 | pdzk1ip1 | -0.130 |
| phlda2 | 0.464 | fbp1b | -0.129 |
| glsb | 0.463 | slc26a1 | -0.126 |
| plcd3a | 0.460 | fbl | -0.126 |
| muc5e | 0.454 | si:ch73-281n10.2 | -0.125 |
| BX000438.2 | 0.452 | nucks1a | -0.125 |
| hhla2a.1 | 0.451 | snrpe | -0.123 |
| tmem176l.2 | 0.451 | hmgb1b | -0.123 |
| si:ch211-153b23.4 | 0.449 | si:ch211-288g17.3 | -0.122 |
| tmem176l.4 | 0.449 | snrnp70 | -0.122 |
| nkpd1 | 0.448 | hmgb2b | -0.122 |
| znf385b | 0.447 | eny2 | -0.122 |
| slc7a1 | 0.445 | hnrnpa0a | -0.119 |
| itga6l | 0.441 | top1l | -0.119 |
| itpk1a | 0.439 | upb1 | -0.119 |