"ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 5"
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 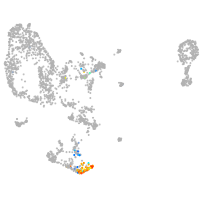 |
neural |
spinal cord / glia |
eye |
taste / olfactory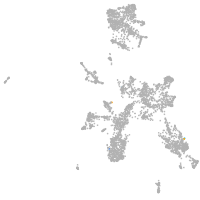 |
otic / lateral line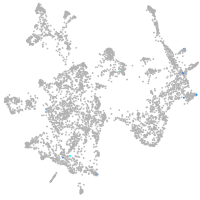 |
epidermis |
periderm |
fin |
ionocytes / mucous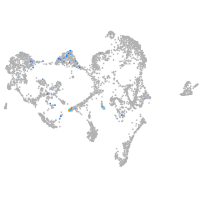 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| atp1a1a.3 | 0.164 | tuba1c | -0.029 |
| bsnd | 0.147 | atp6v0cb | -0.028 |
| trim35-12 | 0.142 | gpm6aa | -0.028 |
| slc12a10.3 | 0.142 | marcksl1a | -0.028 |
| rhcgb | 0.131 | gpm6ab | -0.026 |
| clcnk | 0.130 | rtn1a | -0.026 |
| etf1a | 0.127 | rnasekb | -0.024 |
| si:ch73-359m17.9 | 0.116 | atpv0e2 | -0.023 |
| si:ch73-14h1.2 | 0.104 | ckbb | -0.023 |
| c1qtnf9 | 0.093 | nova2 | -0.023 |
| sstr5 | 0.093 | stmn1b | -0.023 |
| tmem72 | 0.088 | cspg5a | -0.022 |
| mal2 | 0.087 | elavl3 | -0.022 |
| si:dkey-39a18.1 | 0.086 | rtn1b | -0.022 |
| gpr4 | 0.085 | sncb | -0.022 |
| kcnj1a.1 | 0.084 | fam168a | -0.021 |
| si:dkey-96g2.1 | 0.082 | gng3 | -0.021 |
| slc12a10.2 | 0.081 | sypb | -0.021 |
| atp1a1a.2 | 0.078 | tubb5 | -0.021 |
| ptprjb.1 | 0.078 | vamp2 | -0.021 |
| prlrb | 0.077 | calm1a | -0.021 |
| gucy2c | 0.073 | atp1a3a | -0.020 |
| emx1 | 0.070 | celf2 | -0.020 |
| clcn2c | 0.069 | stx1b | -0.020 |
| sgk2a | 0.069 | cadm3 | -0.019 |
| si:dkey-189h5.6 | 0.069 | elavl4 | -0.019 |
| prlra | 0.067 | gapdhs | -0.019 |
| si:dkey-36i7.3 | 0.067 | mdkb | -0.019 |
| exoc3l2b | 0.066 | nsg2 | -0.019 |
| malb | 0.066 | olfm1b | -0.019 |
| si:dkey-33i11.4 | 0.066 | si:dkeyp-75h12.5 | -0.019 |
| foxi3b | 0.065 | cpe | -0.018 |
| nkpd1 | 0.065 | epb41a | -0.018 |
| slc12a3 | 0.065 | hbbe1.3 | -0.018 |
| slc9a3.1 | 0.064 | ndrg4 | -0.018 |




