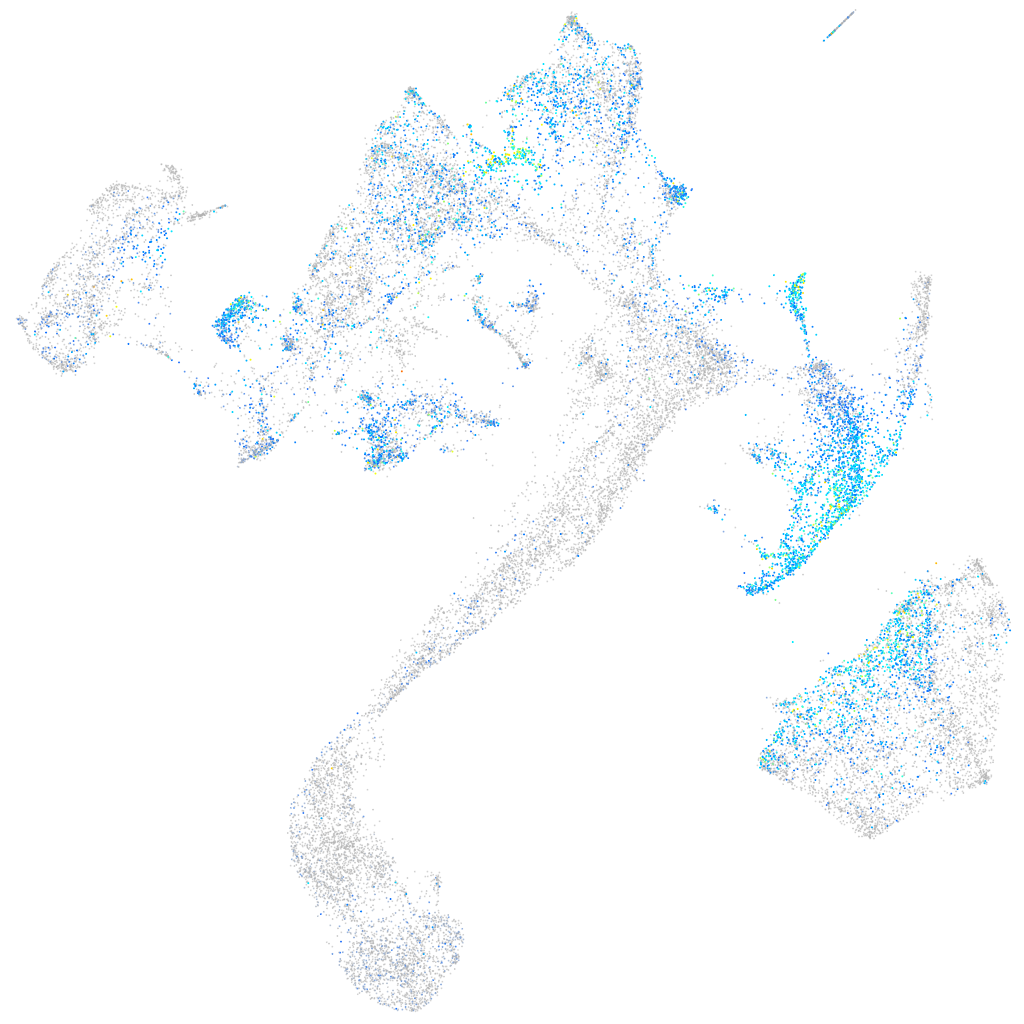
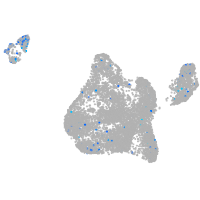
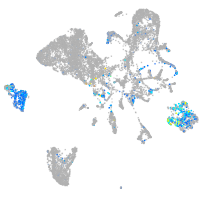
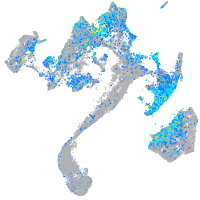
retinoic acid receptor gamma a
ZFIN 
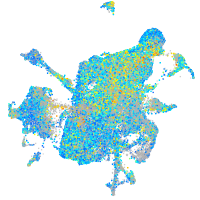
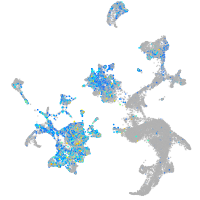
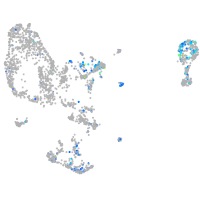
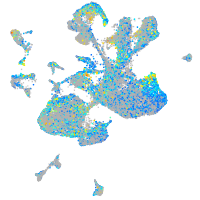
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| itm2cb | 0.421 | ttn.2 | -0.267 |
| hes6 | 0.357 | ttn.1 | -0.266 |
| hoxb10a | 0.343 | actc1b | -0.265 |
| her12 | 0.343 | aldoab | -0.242 |
| her7 | 0.338 | pabpc4 | -0.240 |
| hoxd12a | 0.337 | tmem38a | -0.234 |
| msgn1 | 0.334 | acta1b | -0.234 |
| her1 | 0.334 | ak1 | -0.234 |
| hoxd10a | 0.324 | tpma | -0.233 |
| hoxa13b | 0.320 | atp2a1 | -0.232 |
| hoxc13a | 0.311 | mybphb | -0.231 |
| tbx16l | 0.308 | klhl41b | -0.230 |
| ptmab | 0.307 | gatm | -0.229 |
| nid2a | 0.301 | ckmb | -0.228 |
| hoxc13b | 0.298 | tnnc2 | -0.227 |
| BX005254.3 | 0.298 | ldb3a | -0.227 |
| apoc1 | 0.296 | gapdh | -0.227 |
| fzd10 | 0.293 | srl | -0.224 |
| hoxb7a | 0.292 | ckma | -0.223 |
| wnt5b | 0.290 | zgc:101853 | -0.221 |
| greb1 | 0.288 | gamt | -0.221 |
| hoxa11a | 0.287 | fxr2 | -0.221 |
| zgc:162939 | 0.284 | CABZ01078594.1 | -0.216 |
| cx43.4 | 0.281 | neb | -0.215 |
| hoxa10b | 0.281 | txlnbb | -0.214 |
| si:ch211-222l21.1 | 0.280 | myl1 | -0.211 |
| sall4 | 0.280 | eno1a | -0.211 |
| hsp90ab1 | 0.276 | rbfox1l | -0.211 |
| gdf11 | 0.275 | cox17 | -0.210 |
| vox | 0.275 | desma | -0.210 |
| phgdh | 0.270 | mylpfa | -0.208 |
| raraa | 0.269 | CABZ01072309.1 | -0.207 |
| ephb3 | 0.266 | ldb3b | -0.205 |
| hoxa11b | 0.266 | actn3a | -0.204 |
| tob1a | 0.262 | zgc:92518 | -0.204 |