"lecithin retinol acyltransferase b, tandem duplicate 2"
ZFIN 
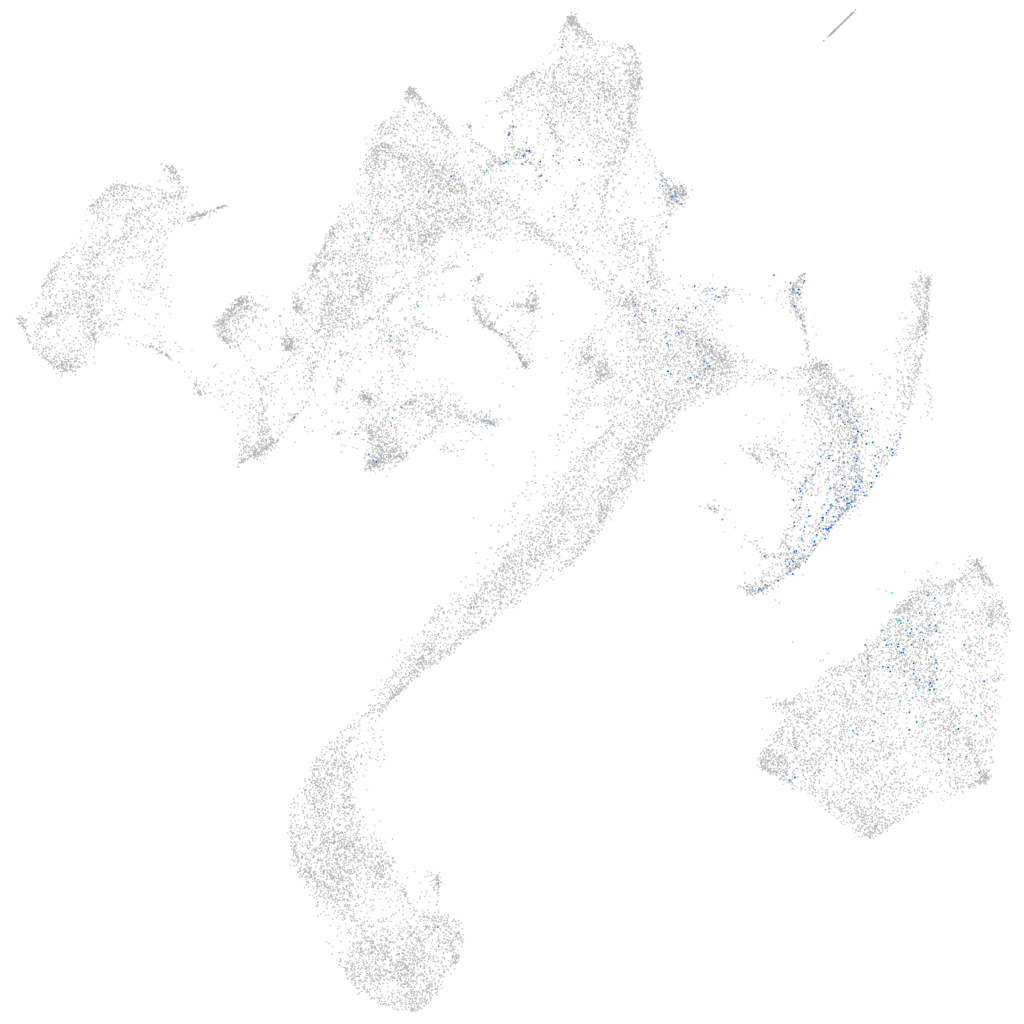
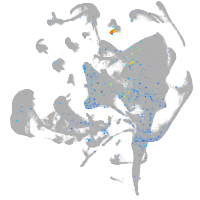
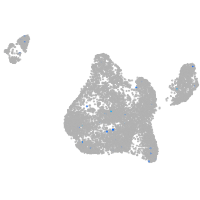
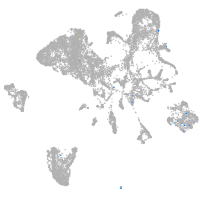
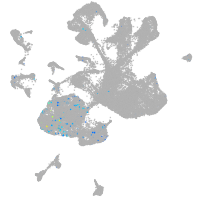
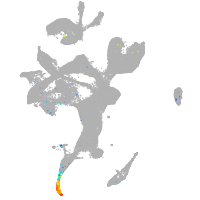
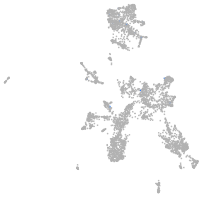
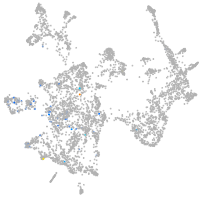
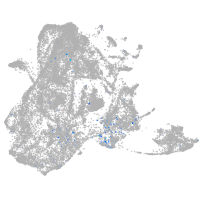
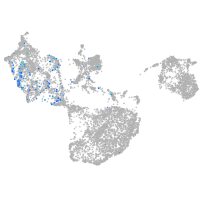
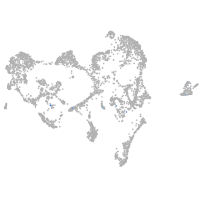
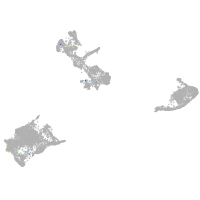
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| itm2cb | 0.163 | fabp3 | -0.084 |
| msgn1 | 0.155 | actc1b | -0.078 |
| her12 | 0.151 | ttn.2 | -0.074 |
| tbx16l | 0.143 | pabpc4 | -0.068 |
| hoxb10a | 0.139 | eef1da | -0.068 |
| fzd10 | 0.139 | klhl41b | -0.067 |
| vox | 0.138 | tmem38a | -0.067 |
| her7 | 0.136 | aldoab | -0.065 |
| hes6 | 0.136 | atp2a1 | -0.065 |
| hoxd10a | 0.129 | ckmb | -0.063 |
| her1 | 0.124 | ckma | -0.063 |
| cx43.4 | 0.124 | ak1 | -0.063 |
| rarga | 0.123 | ttn.1 | -0.063 |
| tob1a | 0.121 | desma | -0.062 |
| sall4 | 0.115 | cox17 | -0.062 |
| nid2a | 0.113 | acta1b | -0.062 |
| cdx4 | 0.113 | acta1a | -0.062 |
| tbx16 | 0.111 | ldb3a | -0.061 |
| hoxd12a | 0.110 | srl | -0.061 |
| hoxb7a | 0.108 | gatm | -0.061 |
| greb1 | 0.108 | mybphb | -0.061 |
| tnfrsf9a | 0.106 | tnnc2 | -0.061 |
| rbm38 | 0.105 | tpma | -0.061 |
| thbs2a | 0.104 | eno1a | -0.060 |
| CABZ01043826.1 | 0.104 | neb | -0.060 |
| inka1b | 0.098 | mylpfa | -0.060 |
| hoxd11a | 0.097 | nexn | -0.060 |
| cyp26a1 | 0.097 | zgc:101853 | -0.060 |
| hoxa10b | 0.096 | gamt | -0.059 |
| hoxa13b | 0.096 | gapdh | -0.058 |
| apoc1 | 0.095 | myl1 | -0.058 |
| dlc | 0.094 | CABZ01078594.1 | -0.058 |
| BX005254.3 | 0.094 | eif4a1b | -0.058 |
| pcdh8 | 0.094 | plecb | -0.057 |
| hoxc11a | 0.093 | FQ323156.1 | -0.057 |