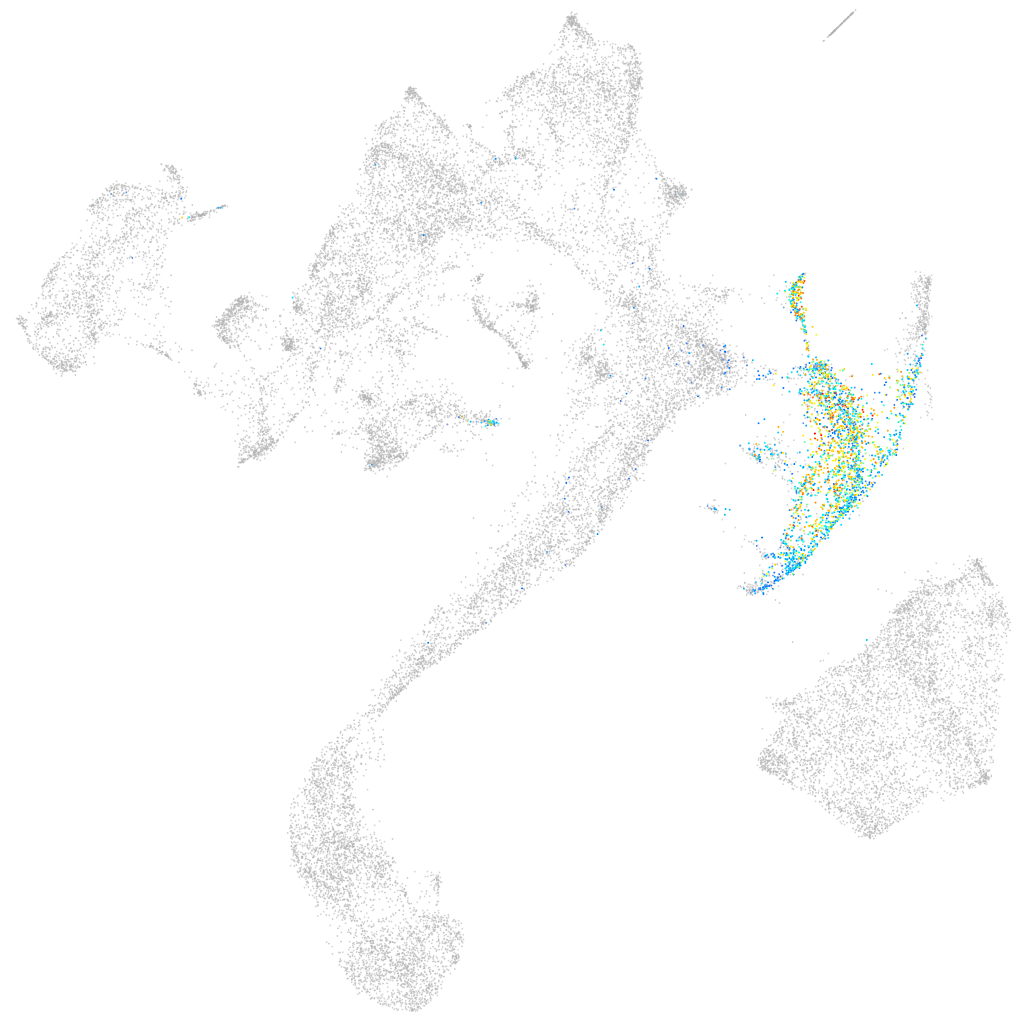
hairy-related 1
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pcdh8 | 0.662 | fabp3 | -0.248 |
| her7 | 0.638 | sparc | -0.224 |
| itm2cb | 0.586 | actc1b | -0.204 |
| hoxb10a | 0.562 | bhmt | -0.186 |
| hoxd10a | 0.555 | pabpc4 | -0.183 |
| her12 | 0.524 | vwde | -0.179 |
| ephb3 | 0.507 | ttn.2 | -0.176 |
| ripply2 | 0.492 | ckma | -0.176 |
| msgn1 | 0.491 | ak1 | -0.176 |
| LO016987.2 | 0.468 | atp2a1 | -0.174 |
| BX005254.3 | 0.468 | ckmb | -0.174 |
| nid2a | 0.467 | eno1a | -0.173 |
| dlc | 0.464 | rplp2 | -0.173 |
| tbx6 | 0.456 | tmem38a | -0.171 |
| apoc1 | 0.455 | gapdh | -0.169 |
| her11 | 0.450 | eef1da | -0.167 |
| hes6 | 0.445 | col1a2 | -0.167 |
| tbx16l | 0.444 | mylpfa | -0.166 |
| fn1b | 0.402 | si:dkey-16p21.8 | -0.165 |
| greb1 | 0.395 | aldoab | -0.164 |
| tspan7 | 0.385 | FQ323156.1 | -0.163 |
| hoxa11a | 0.384 | gamt | -0.158 |
| hoxd12a | 0.383 | acta1b | -0.156 |
| draxin | 0.378 | srl | -0.156 |
| myf5 | 0.377 | tnnc2 | -0.154 |
| hoxa11b | 0.377 | neb | -0.153 |
| sall4 | 0.373 | col1a1a | -0.153 |
| XLOC-042222 | 0.372 | cox17 | -0.153 |
| gnaia | 0.365 | myl1 | -0.152 |
| hoxb7a | 0.362 | pvalb1 | -0.152 |
| phc2a | 0.354 | CABZ01078594.1 | -0.152 |
| hoxa10b | 0.352 | ldb3a | -0.151 |
| fbln1 | 0.351 | pvalb2 | -0.150 |
| inka1b | 0.350 | cav3 | -0.150 |
| hoxd11a | 0.348 | cd81a | -0.149 |