FEZ family zinc finger 1
ZFIN 
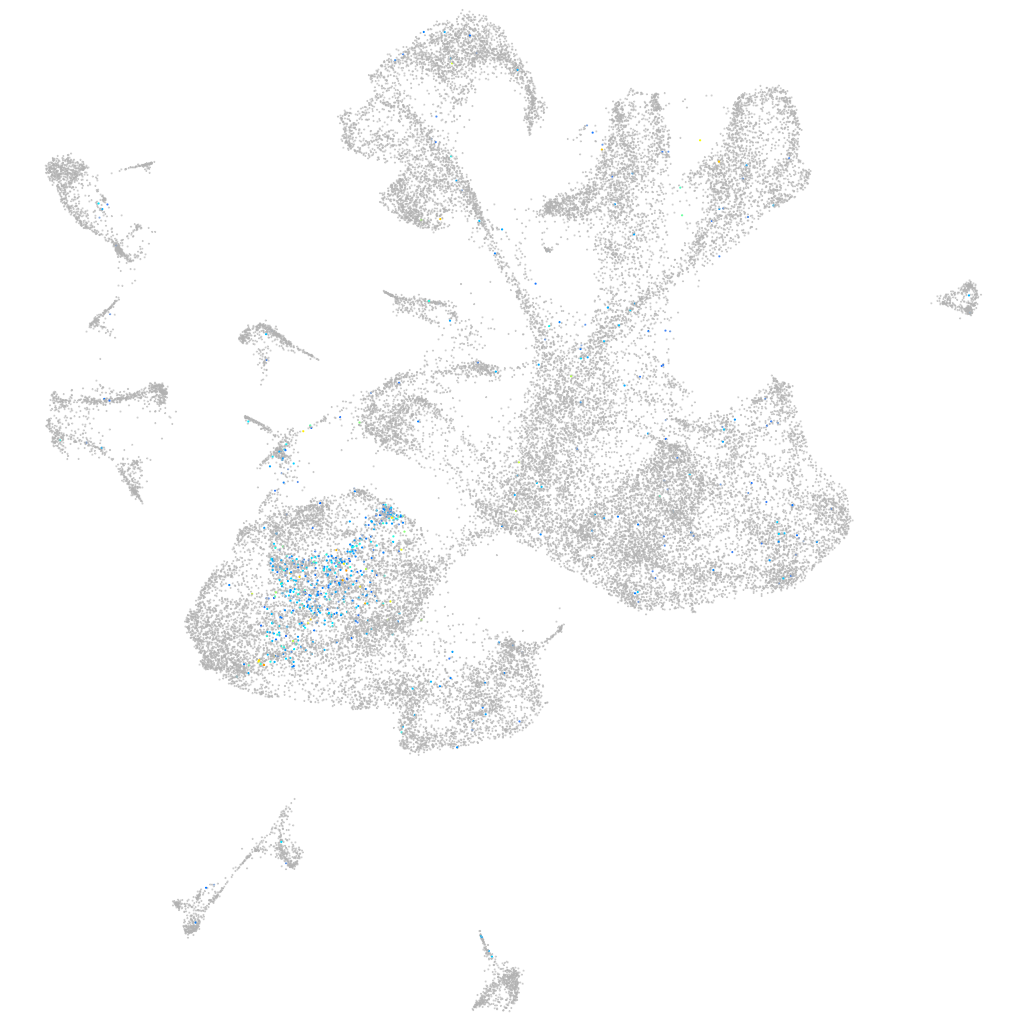
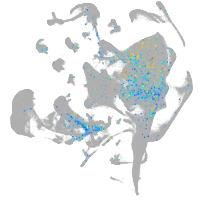
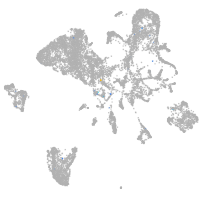
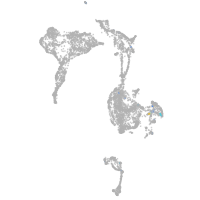
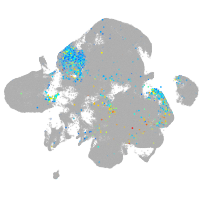
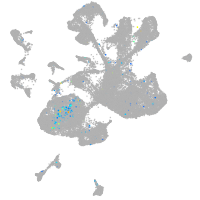
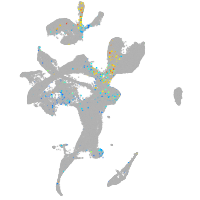
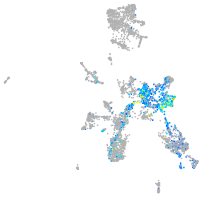
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| foxg1a | 0.279 | hoxb3a | -0.066 |
| vax1 | 0.159 | meis1b | -0.062 |
| six3a | 0.151 | hoxb9a | -0.060 |
| fezf2 | 0.140 | hoxc3a | -0.057 |
| fgfrl1a | 0.137 | si:ch73-265d7.2 | -0.054 |
| lhx2b | 0.128 | hoxb8a | -0.053 |
| CR751602.2 | 0.127 | hoxc9a | -0.049 |
| hs3st1l2 | 0.124 | hoxd3a | -0.048 |
| CABZ01030107.1 | 0.122 | hoxb1b | -0.046 |
| nr2e1 | 0.122 | hoxa9a | -0.045 |
| cadps2 | 0.121 | hoxa3a | -0.044 |
| zgc:165461 | 0.116 | hoxb7a | -0.043 |
| si:ch211-156b7.4 | 0.105 | nr2f6b | -0.043 |
| lix1 | 0.105 | pax3a | -0.042 |
| lrig1 | 0.099 | si:ch73-386h18.1 | -0.042 |
| slc1a3b | 0.099 | marcksl1b | -0.042 |
| six3b | 0.098 | cdkn1ca | -0.042 |
| six6a | 0.097 | calm2a | -0.041 |
| gsx2 | 0.094 | tmeff1b | -0.041 |
| CU467822.1 | 0.090 | hoxa9b | -0.041 |
| nkx2.1 | 0.088 | serinc5 | -0.041 |
| efnb1 | 0.086 | hoxc8a | -0.040 |
| podxl | 0.084 | epha4b | -0.040 |
| sfrp5 | 0.083 | hoxd9a | -0.040 |
| celf2 | 0.083 | ldlrad2 | -0.039 |
| eomesa | 0.082 | sb:cb81 | -0.039 |
| ppap2d | 0.082 | vim | -0.039 |
| emx3 | 0.080 | elavl3 | -0.039 |
| fgfr2 | 0.078 | hoxc6b | -0.038 |
| foxd2 | 0.077 | hoxb10a | -0.038 |
| eno4 | 0.077 | hoxc1a | -0.038 |
| BX005454.1 | 0.074 | snap25a | -0.038 |
| znf296 | 0.074 | si:ch211-57n23.4 | -0.038 |
| dmrta2 | 0.074 | tubb5 | -0.037 |
| cd82a | 0.072 | hoxa10b | -0.037 |