"retinoic acid receptor, gamma b"
ZFIN 
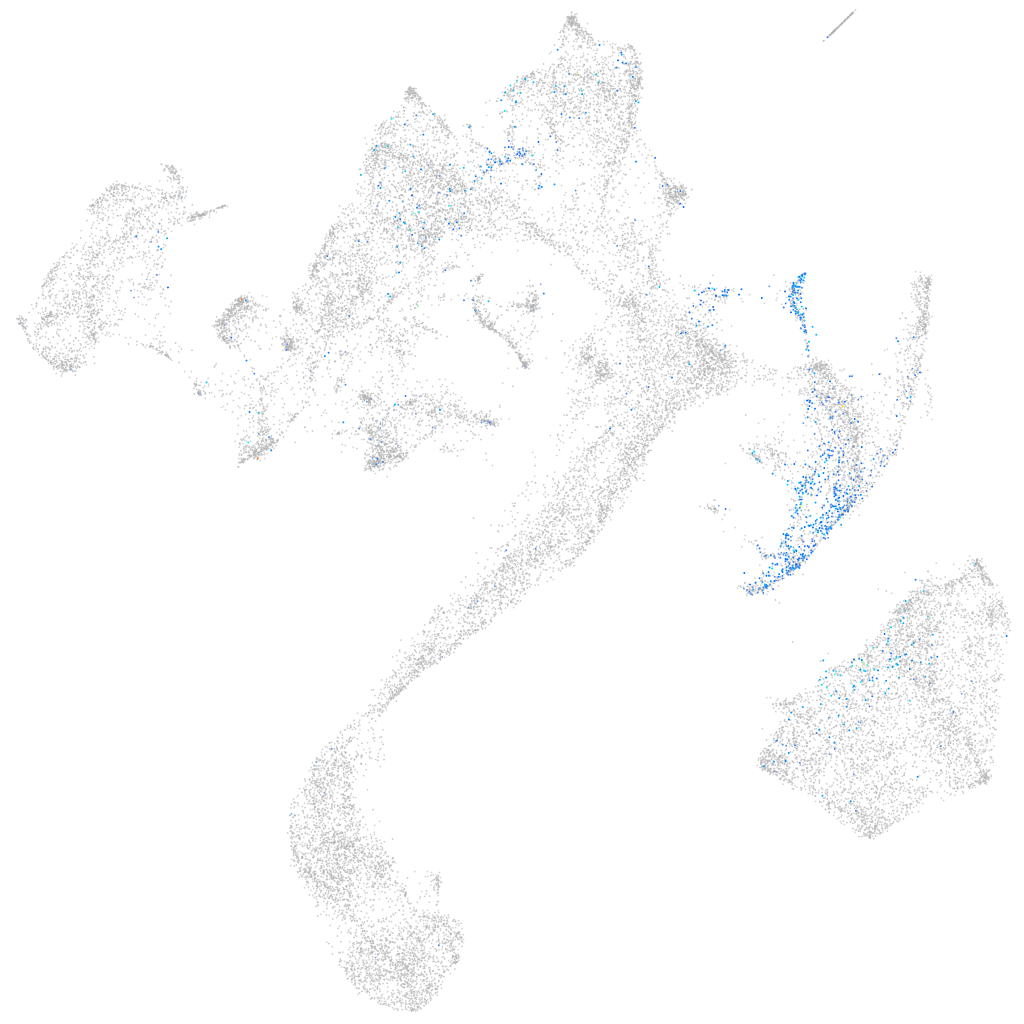
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hoxd12a | 0.271 | ttn.2 | -0.115 |
| hoxa13b | 0.260 | actc1b | -0.115 |
| itm2cb | 0.260 | ttn.1 | -0.112 |
| her1 | 0.243 | aldoab | -0.102 |
| hoxd10a | 0.235 | ak1 | -0.101 |
| rarga | 0.234 | atp2a1 | -0.098 |
| her7 | 0.226 | ckmb | -0.098 |
| gdf11 | 0.220 | gapdh | -0.098 |
| hoxc13b | 0.218 | pabpc4 | -0.097 |
| hoxc13a | 0.215 | ckma | -0.097 |
| hoxb10a | 0.215 | klhl41b | -0.097 |
| her12 | 0.214 | tmem38a | -0.096 |
| nid2a | 0.208 | eno1a | -0.096 |
| hes6 | 0.203 | cox17 | -0.095 |
| hoxa11a | 0.198 | tpma | -0.095 |
| cyp26a1 | 0.198 | tnnc2 | -0.094 |
| hoxc11a | 0.188 | acta1b | -0.094 |
| tagln3b | 0.187 | gamt | -0.093 |
| fbln1 | 0.187 | mybphb | -0.092 |
| hoxa13a | 0.186 | gatm | -0.092 |
| fgf3 | 0.183 | fxr2 | -0.091 |
| zgc:162939 | 0.183 | srl | -0.091 |
| msgn1 | 0.183 | desma | -0.091 |
| thbs2a | 0.181 | mylpfa | -0.090 |
| hoxd11a | 0.177 | neb | -0.090 |
| sall4 | 0.175 | fabp3 | -0.090 |
| hoxc11b | 0.174 | ldb3a | -0.090 |
| hoxc12b | 0.174 | tpi1b | -0.089 |
| apoc1 | 0.173 | CABZ01078594.1 | -0.089 |
| tbx16l | 0.172 | myl1 | -0.087 |
| vox | 0.172 | zgc:101853 | -0.087 |
| greb1 | 0.172 | txlnbb | -0.087 |
| hoxc10a | 0.170 | acta1a | -0.086 |
| fzd10 | 0.167 | zgc:92518 | -0.086 |
| ephb3 | 0.166 | ldb3b | -0.086 |