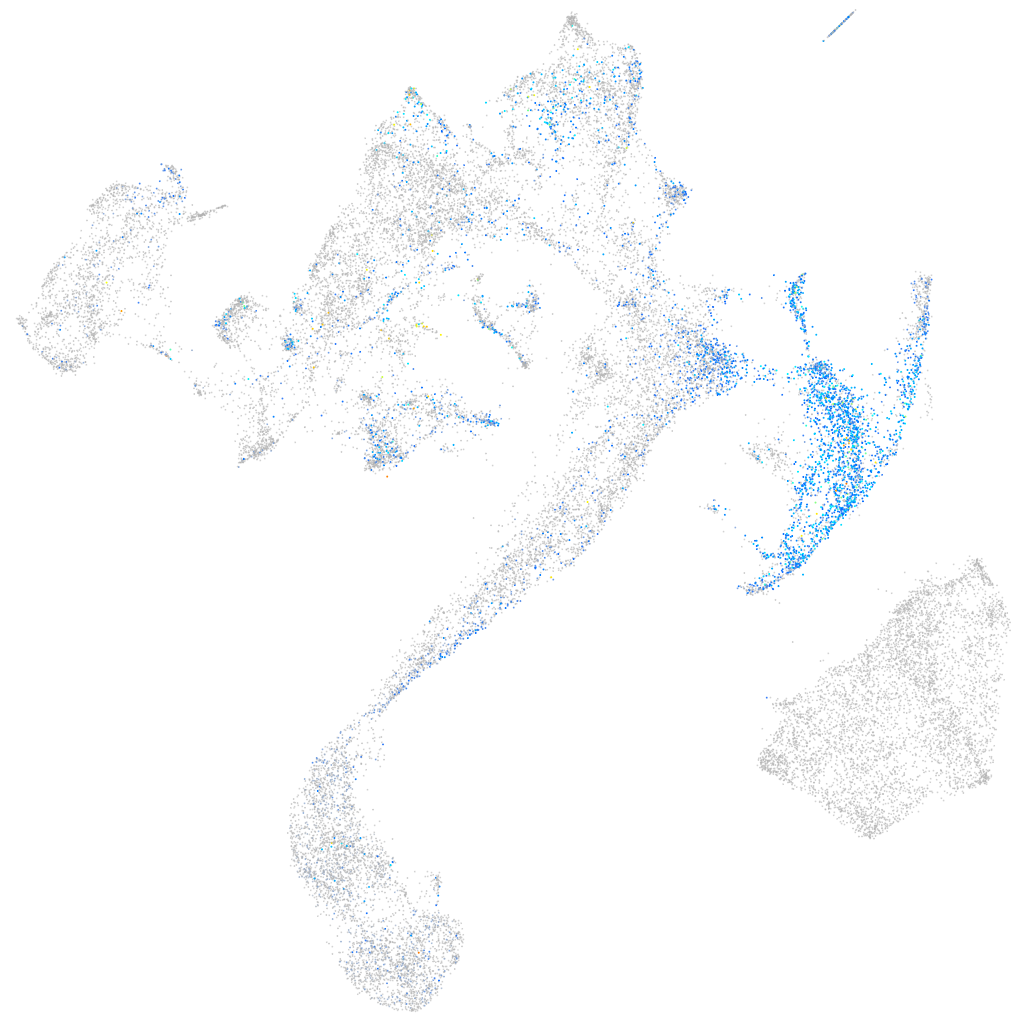
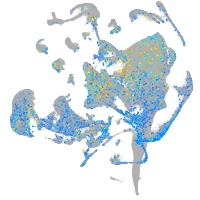
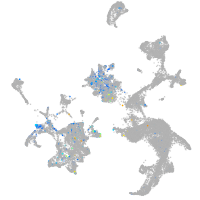
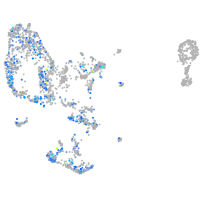
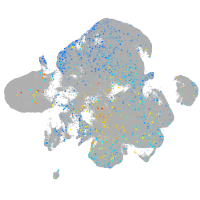
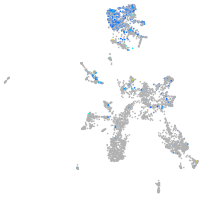
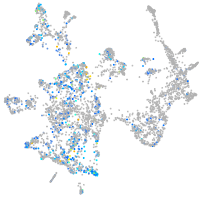
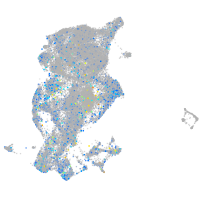
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pcdh8 | 0.476 | fabp3 | -0.137 |
| her1 | 0.468 | actc1b | -0.137 |
| ephb3 | 0.462 | ckmb | -0.134 |
| hoxb10a | 0.371 | si:ch211-152c2.3 | -0.133 |
| hoxd10a | 0.356 | ckma | -0.132 |
| itm2cb | 0.350 | atp2a1 | -0.131 |
| nid2a | 0.324 | si:dkey-16p21.8 | -0.131 |
| her7 | 0.318 | pabpc4 | -0.131 |
| fn1b | 0.316 | gapdh | -0.129 |
| draxin | 0.304 | ak1 | -0.128 |
| her12 | 0.303 | tmem38a | -0.122 |
| tbx6 | 0.303 | stm | -0.122 |
| XLOC-042222 | 0.295 | cox7c | -0.121 |
| tspan7 | 0.287 | nme2b.2 | -0.119 |
| sall4 | 0.286 | gamt | -0.119 |
| BX005254.3 | 0.279 | aldoab | -0.119 |
| hnrnpa0l | 0.277 | FQ323156.1 | -0.115 |
| greb1 | 0.274 | rplp2 | -0.113 |
| ripply2 | 0.269 | neb | -0.112 |
| hoxa11b | 0.262 | glud1b | -0.112 |
| dlc | 0.256 | mylpfa | -0.112 |
| hoxa11a | 0.254 | COX7A2 | -0.112 |
| fbln1 | 0.252 | rps18 | -0.112 |
| apoc1 | 0.249 | eno1a | -0.112 |
| phc2a | 0.248 | si:ch73-367p23.2 | -0.111 |
| myf5 | 0.247 | ank1a | -0.111 |
| igsf9a | 0.245 | tnnc2 | -0.111 |
| zbtb16a | 0.243 | atp5md | -0.110 |
| hoxd11a | 0.241 | ldb3b | -0.110 |
| hoxa10b | 0.241 | tnnt3a | -0.110 |
| has2 | 0.240 | srl | -0.110 |
| inka1b | 0.240 | cav3 | -0.109 |
| adgrl2a | 0.240 | pgam2 | -0.109 |
| hoxc11a | 0.240 | tmod4 | -0.109 |
| msgn1 | 0.239 | CABZ01078594.1 | -0.109 |