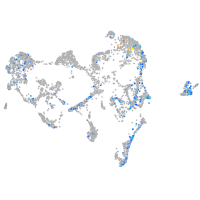
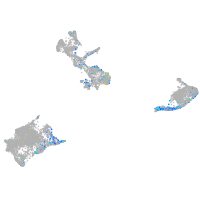
homeobox C3a
ZFIN 
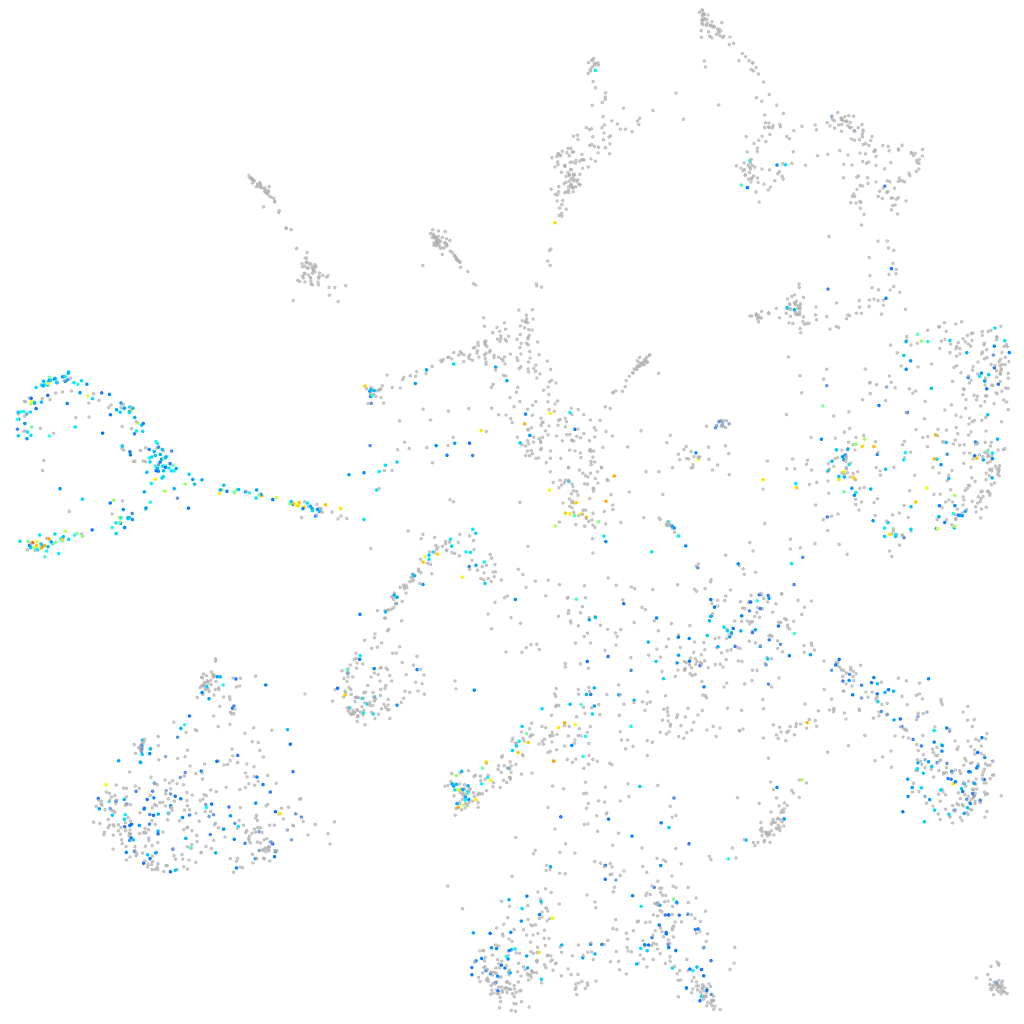
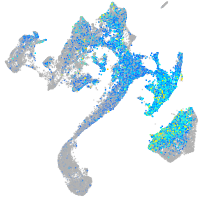
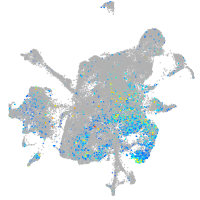
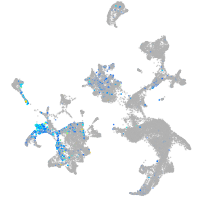
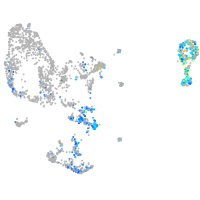
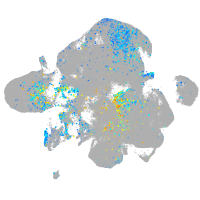
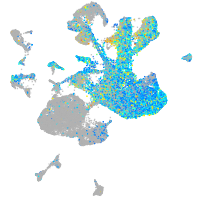
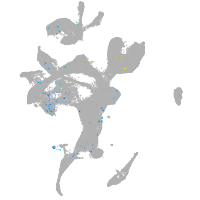
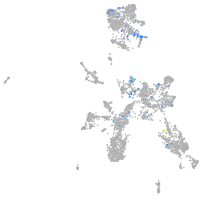
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hoxc8a | 0.395 | notch3 | -0.128 |
| ldha | 0.363 | bambia | -0.121 |
| pkma | 0.345 | rbpms2b | -0.116 |
| eno3 | 0.320 | ednraa | -0.115 |
| aldocb | 0.318 | tmsb4x | -0.114 |
| inhbaa | 0.308 | rplp2 | -0.109 |
| loxl5b | 0.305 | krt94 | -0.108 |
| hoxb8a | 0.305 | zeb2a | -0.107 |
| pgk1 | 0.303 | rasl12 | -0.107 |
| barx1 | 0.302 | akap12b | -0.107 |
| gapdhs | 0.293 | COLEC10 | -0.106 |
| pgam1a | 0.279 | foxc1a | -0.105 |
| eno1a | 0.273 | mmel1 | -0.103 |
| pfkpa | 0.269 | foxc1b | -0.100 |
| tpi1b | 0.268 | si:dkey-238c7.16 | -0.100 |
| pgp | 0.263 | c1qtnf6a | -0.100 |
| slc16a3 | 0.262 | twist1b | -0.099 |
| fbp2 | 0.257 | nid1a | -0.096 |
| gpia | 0.255 | si:dkey-57k2.6 | -0.096 |
| wnt11r | 0.249 | colec11 | -0.095 |
| hk1 | 0.243 | vim | -0.095 |
| bgna | 0.238 | lsp1 | -0.094 |
| angptl5 | 0.233 | rhag | -0.094 |
| hoxc1a | 0.232 | gata5 | -0.093 |
| slc2a1b | 0.226 | junba | -0.092 |
| namptb | 0.224 | smad9 | -0.092 |
| pitx1 | 0.223 | tnfsf12 | -0.090 |
| alcama | 0.218 | ecm1b | -0.088 |
| mgll | 0.215 | twist1a | -0.085 |
| cx43 | 0.211 | dub | -0.083 |
| grem2b | 0.208 | LO016987.2 | -0.083 |
| isl2b | 0.203 | fabp11a | -0.082 |
| pthlha | 0.193 | pcolcea | -0.081 |
| si:dkey-44g17.6 | 0.188 | sfrp5 | -0.081 |
| sirt5 | 0.188 | cyp3c1 | -0.080 |