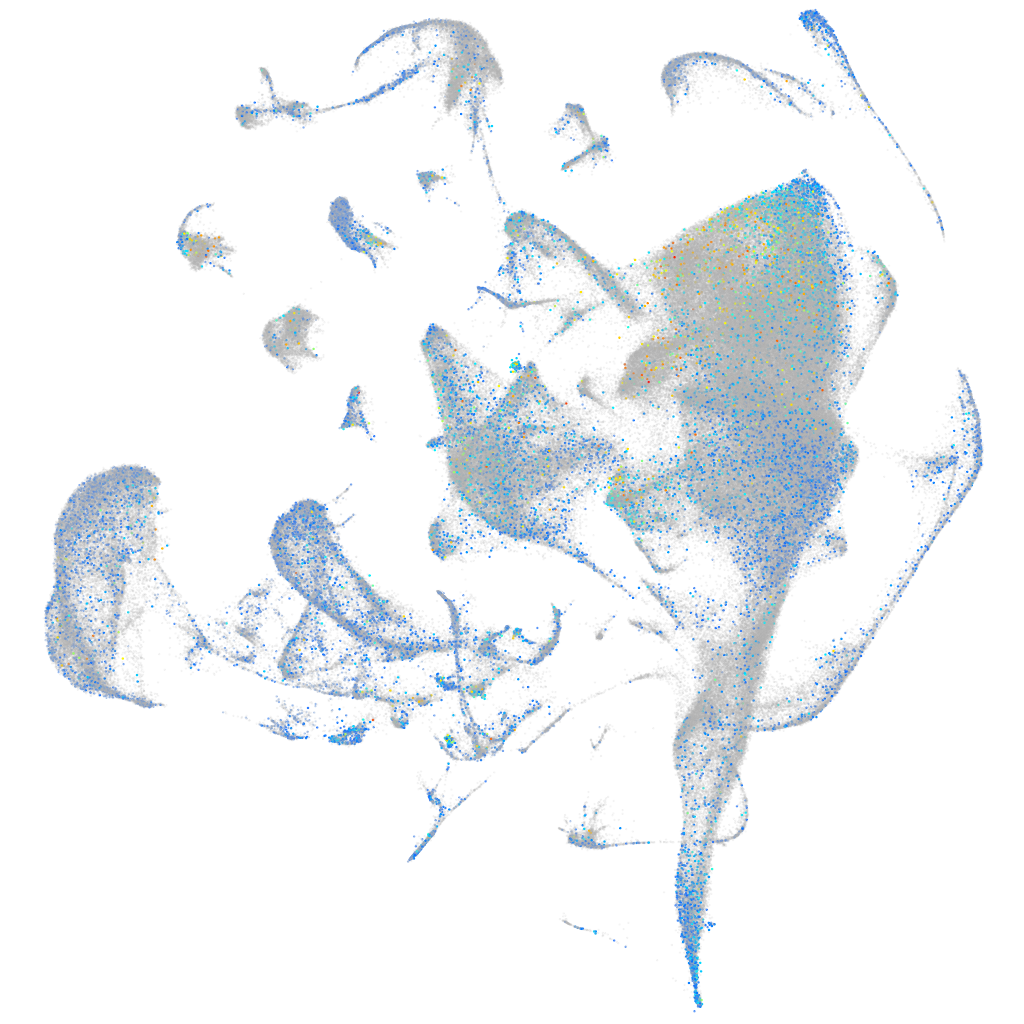sirtuin 5
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| eno3 | 0.071 | dla | -0.034 |
| ldhba | 0.070 | dlb | -0.033 |
| atp5if1b | 0.068 | LOC798783 | -0.033 |
| atp5mc1 | 0.067 | si:ch211-137a8.4 | -0.032 |
| pgk1 | 0.067 | si:dkey-56m19.5 | -0.032 |
| tpi1b | 0.066 | nova2 | -0.031 |
| chchd10 | 0.064 | gadd45gb.1 | -0.030 |
| ppifb | 0.063 | notch1a | -0.030 |
| sod2 | 0.062 | sox11b | -0.030 |
| gpia | 0.060 | XLOC-003689 | -0.030 |
| suclg1 | 0.060 | hes6 | -0.029 |
| pgam1a | 0.059 | si:ch73-21g5.7 | -0.029 |
| pkma | 0.057 | tp53inp2 | -0.029 |
| atp5meb | 0.056 | XLOC-003692 | -0.029 |
| sod1 | 0.055 | abhd6a | -0.029 |
| ddt | 0.054 | neurod4 | -0.028 |
| sdhdb | 0.054 | chd4a | -0.027 |
| STMP1 | 0.054 | marcksb | -0.027 |
| atp5f1c | 0.052 | pou3f1 | -0.027 |
| cox17 | 0.052 | si:ch211-57n23.4 | -0.027 |
| glo1 | 0.052 | scrt2 | -0.026 |
| acadm | 0.051 | her15.1 | -0.026 |
| dlst | 0.051 | her4.2 | -0.026 |
| gapdhs | 0.051 | cdkn1ca | -0.025 |
| mdh2 | 0.051 | ebf2 | -0.025 |
| mpc1 | 0.051 | LOC101882472 | -0.025 |
| aldh6a1 | 0.050 | rtca | -0.025 |
| atp5f1d | 0.050 | CU634008.1 | -0.025 |
| eef1da | 0.050 | FO082781.1 | -0.024 |
| eno1a | 0.050 | hmgb1b | -0.024 |
| fdx1 | 0.050 | lfng | -0.024 |
| glud1b | 0.050 | XLOC-003690 | -0.024 |
| got2b | 0.050 | hbbe1.3 | -0.023 |
| minos1 | 0.050 | her13 | -0.023 |
| prdx3 | 0.050 | hnrnpa1a | -0.023 |