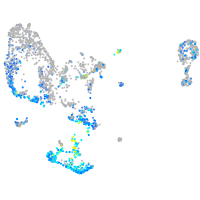
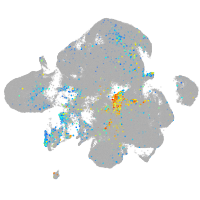
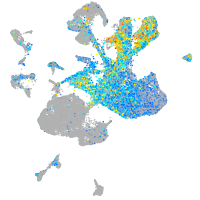
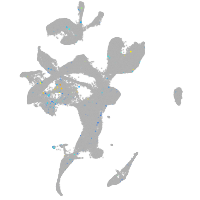
homeobox B8a
ZFIN 
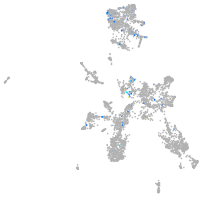
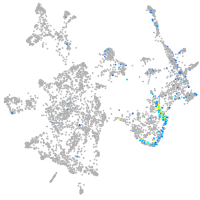
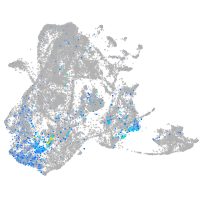
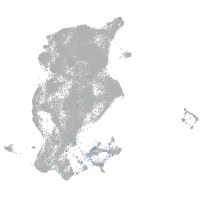
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hoxc8a | 0.364 | tmsb4x | -0.166 |
| hoxc3a | 0.305 | krt94 | -0.147 |
| ldha | 0.261 | ifitm1 | -0.116 |
| loxl5b | 0.239 | mcl1b | -0.113 |
| pkma | 0.238 | COLEC10 | -0.109 |
| inhbaa | 0.231 | gata5 | -0.106 |
| hoxc6a | 0.228 | ephb3 | -0.105 |
| aldocb | 0.223 | pcolcea | -0.104 |
| barx1 | 0.219 | rbpms2b | -0.100 |
| eno3 | 0.207 | mmel1 | -0.099 |
| hk1 | 0.205 | foxc1b | -0.099 |
| hoxb6a | 0.204 | si:dkey-57k2.6 | -0.095 |
| si:dkey-1c11.1 | 0.200 | rasl12 | -0.093 |
| gapdhs | 0.199 | smad9 | -0.092 |
| hoxb9a | 0.198 | twist1b | -0.092 |
| ntn5 | 0.197 | cav1 | -0.091 |
| bgna | 0.195 | ecm1b | -0.090 |
| meis2b | 0.189 | vim | -0.090 |
| pbx3b | 0.186 | bambia | -0.088 |
| eno1a | 0.183 | cd151 | -0.087 |
| pgk1 | 0.181 | colec11 | -0.086 |
| tpi1b | 0.178 | ednraa | -0.086 |
| alcama | 0.177 | si:dkey-238c7.16 | -0.085 |
| asip2b | 0.176 | lsp1 | -0.085 |
| hoxb7a | 0.176 | sh3bgrl2 | -0.085 |
| wnt11r | 0.171 | krt15 | -0.085 |
| hoxc1a | 0.170 | nrp1a | -0.085 |
| pgp | 0.167 | fabp11a | -0.084 |
| sept3 | 0.166 | fosl2 | -0.083 |
| grem2b | 0.164 | cavin1b | -0.082 |
| rdh10a | 0.160 | foxc1a | -0.081 |
| pgam1a | 0.160 | cdh11 | -0.080 |
| foxf1 | 0.160 | LO016987.2 | -0.079 |
| slc2a1b | 0.158 | dub | -0.079 |
| angptl5 | 0.155 | clu | -0.078 |