ADP-ribosylation factor-like 4D
ZFIN 
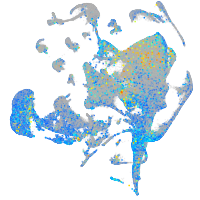
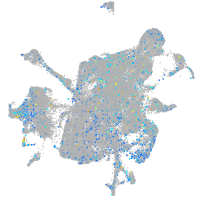
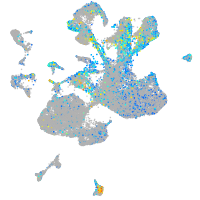
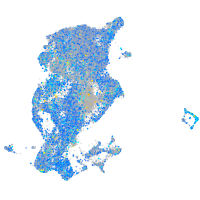
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| vim | 0.107 | gpm6aa | -0.050 |
| hoxc1a | 0.093 | celf2 | -0.049 |
| lin28a | 0.080 | ptmaa | -0.048 |
| dhrs3b | 0.078 | CU467822.1 | -0.047 |
| CABZ01075068.1 | 0.075 | ppiab | -0.039 |
| hoxb5b | 0.070 | CU634008.1 | -0.039 |
| sox32 | 0.069 | tuba1c | -0.037 |
| si:ch1073-284b18.2 | 0.068 | pvalb1 | -0.036 |
| akap12b | 0.065 | cspg5a | -0.036 |
| hoxb1b | 0.063 | marcksl1a | -0.035 |
| hoxd4a | 0.063 | tmsb4x | -0.035 |
| hoxc3a | 0.061 | bcl11ba | -0.035 |
| meis3 | 0.061 | gpm6ab | -0.035 |
| hand2 | 0.059 | nfil3-5 | -0.034 |
| hoxa3a | 0.058 | ckbb | -0.034 |
| hoxb6b | 0.058 | chd4a | -0.034 |
| onecutl | 0.057 | h3f3a | -0.034 |
| hoxc4a | 0.057 | en2b | -0.034 |
| hoxb3a | 0.057 | rtn1b | -0.034 |
| vipb | 0.056 | pvalb2 | -0.033 |
| apoc1 | 0.055 | hbbe1.3 | -0.033 |
| sox17 | 0.054 | stmn1b | -0.032 |
| cdx4 | 0.054 | lhx9 | -0.032 |
| tuba8l3 | 0.054 | CR848047.1 | -0.031 |
| trim71 | 0.053 | actc1b | -0.031 |
| ved | 0.052 | bcl11aa | -0.031 |
| greb1l | 0.052 | cd99l2 | -0.031 |
| nes | 0.052 | rtn1a | -0.031 |
| onecut1 | 0.051 | mir181b-3 | -0.030 |
| si:ch211-215d8.2 | 0.051 | hbae3 | -0.030 |
| arid3b | 0.051 | tcf7l2 | -0.030 |
| hoxb5a | 0.050 | rpl37 | -0.029 |
| gata5 | 0.050 | pax7a | -0.028 |
| apoeb | 0.050 | nova2 | -0.028 |
| zgc:77784 | 0.050 | tubb5 | -0.028 |