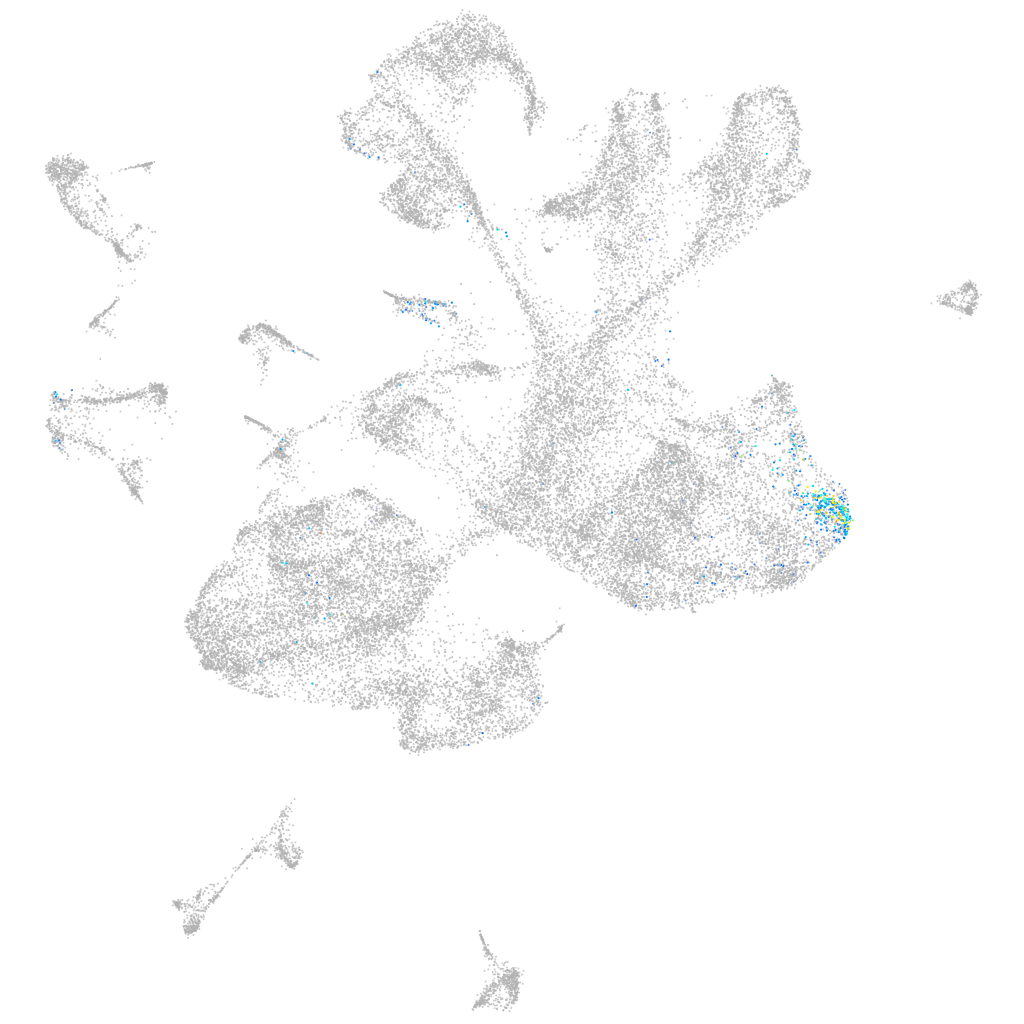
notochord homeobox
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 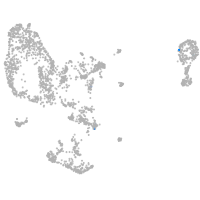 |
neural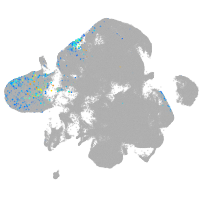 |
spinal cord / glia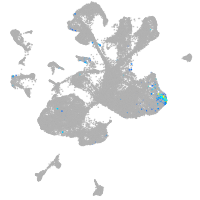 |
eye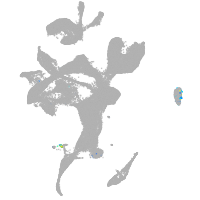 |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sp5l | 0.499 | fabp3 | -0.145 |
| fgf8a | 0.491 | marcksl1b | -0.123 |
| cdx1a | 0.438 | CR383676.1 | -0.107 |
| nkx1.2la | 0.409 | gpm6aa | -0.106 |
| pou5f3 | 0.380 | tmsb4x | -0.096 |
| nradd | 0.376 | nova2 | -0.094 |
| chrd | 0.370 | ckbb | -0.086 |
| lrrc17 | 0.370 | meis1b | -0.084 |
| cxcr4a | 0.346 | rtn1a | -0.081 |
| foxd5 | 0.343 | fabp7a | -0.080 |
| cdx4 | 0.321 | pvalb1 | -0.079 |
| hspb1 | 0.310 | hbbe1.3 | -0.077 |
| apela | 0.306 | pvalb2 | -0.076 |
| tbxta | 0.292 | vim | -0.075 |
| znfl2a | 0.274 | COX3 | -0.073 |
| cpm | 0.247 | actc1b | -0.072 |
| XLOC-042222 | 0.244 | tmsb | -0.070 |
| pnx | 0.244 | hbae3 | -0.070 |
| BX927258.1 | 0.233 | CU467822.1 | -0.069 |
| BX005254.3 | 0.226 | elavl3 | -0.068 |
| notum1a | 0.225 | gapdhs | -0.066 |
| sp5a | 0.224 | zgc:165461 | -0.066 |
| znfl1k | 0.217 | gpm6bb | -0.065 |
| XLOC-001603 | 0.208 | gpm6ab | -0.064 |
| sp9 | 0.207 | stmn1b | -0.064 |
| nid2a | 0.205 | CU634008.1 | -0.063 |
| her3 | 0.205 | hbae1.1 | -0.063 |
| hoxd10a | 0.201 | mdka | -0.063 |
| aebp1 | 0.199 | gfap | -0.062 |
| dnase1l4.1 | 0.198 | atp1a1b | -0.061 |
| hoxd12a | 0.196 | ncam1a | -0.061 |
| BX001014.2 | 0.196 | nr2f1a | -0.061 |
| hoxb10a | 0.195 | rnasekb | -0.061 |
| XLOC-032526 | 0.191 | cspg5a | -0.060 |
| greb1 | 0.190 | ppdpfb | -0.060 |