nuclear factor I/A
ZFIN 
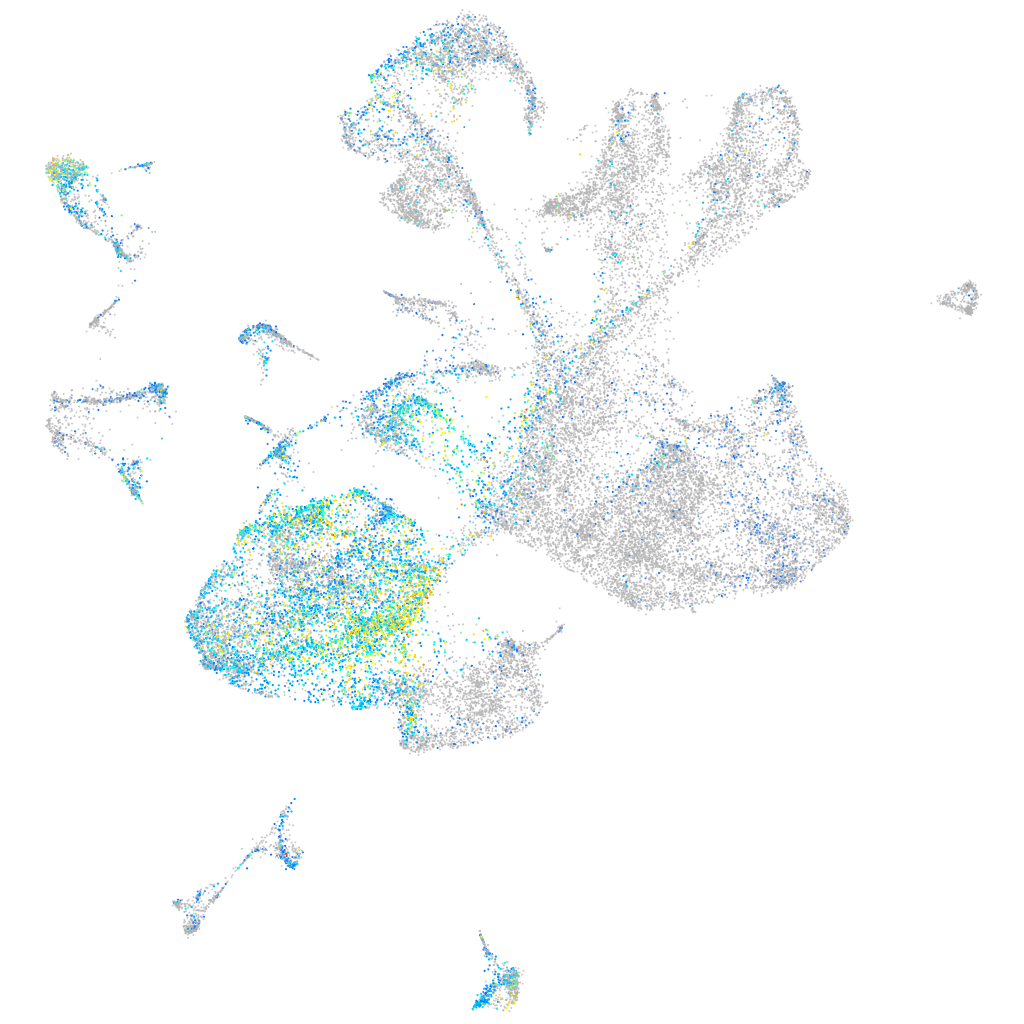
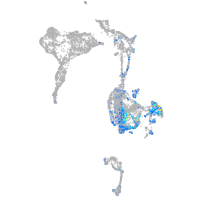
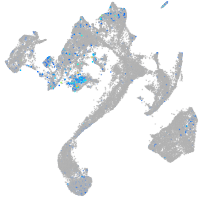
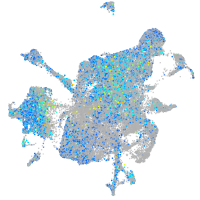
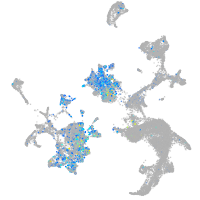
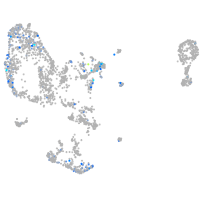
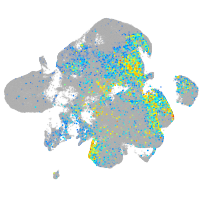
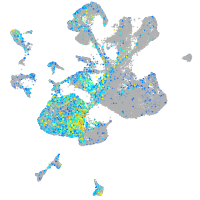
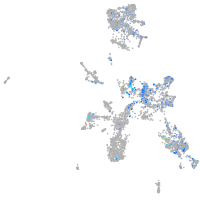
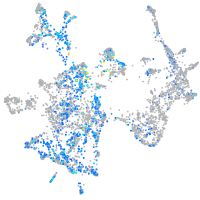
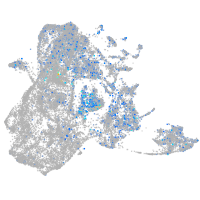
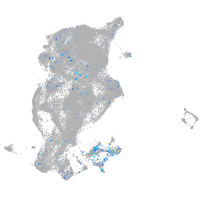
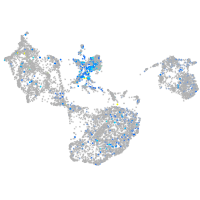
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CR848047.1 | 0.532 | hoxc3a | -0.232 |
| CU467822.1 | 0.459 | si:ch73-386h18.1 | -0.228 |
| fabp7a | 0.437 | hoxb9a | -0.221 |
| nfixb | 0.410 | si:ch211-57n23.4 | -0.219 |
| slc1a2b | 0.366 | ptmab | -0.218 |
| CU634008.1 | 0.337 | CABZ01075068.1 | -0.207 |
| LOC103909284 | 0.330 | tubb2b | -0.204 |
| gpm6bb | 0.327 | slc1a3a | -0.204 |
| masp1 | 0.326 | h3f3a | -0.203 |
| zgc:165461 | 0.287 | si:ch211-222l21.1 | -0.200 |
| ppap2d | 0.285 | hoxb3a | -0.191 |
| ckbb | 0.284 | hmga1a | -0.189 |
| atp1a1b | 0.283 | h3f3d | -0.188 |
| slc1a3b | 0.280 | stmn1a | -0.186 |
| atp1b4 | 0.272 | hoxa9a | -0.177 |
| qki2 | 0.271 | nr2f6b | -0.177 |
| cspg5a | 0.267 | zfhx3 | -0.175 |
| mdkb | 0.266 | hoxb1b | -0.173 |
| cspg5b | 0.265 | hoxd3a | -0.171 |
| tcf4 | 0.264 | zgc:100920 | -0.167 |
| itm2ba | 0.264 | nfyba | -0.166 |
| efhd1 | 0.261 | lin28a | -0.165 |
| ptn | 0.260 | hoxd9a | -0.165 |
| si:ch1073-303k11.2 | 0.258 | hoxb8a | -0.163 |
| chfr | 0.254 | hoxc8a | -0.163 |
| slc6a1b | 0.254 | cx43.4 | -0.162 |
| gpr37l1b | 0.254 | elavl3 | -0.160 |
| hepacama | 0.253 | hoxb10a | -0.159 |
| si:ch73-215f7.1 | 0.251 | hsp90ab1 | -0.158 |
| nck1a | 0.249 | sumo3a | -0.157 |
| cyp26b1 | 0.249 | si:ch211-288g17.3 | -0.156 |
| NPAS3 | 0.244 | hoxc9a | -0.154 |
| sox9a | 0.240 | si:ch73-1a9.3 | -0.153 |
| ptprz1a | 0.238 | mex3b | -0.151 |
| cd82a | 0.235 | alcamb | -0.149 |