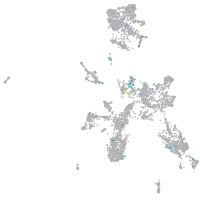
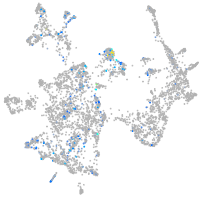
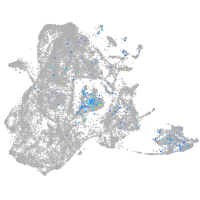
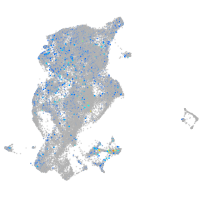
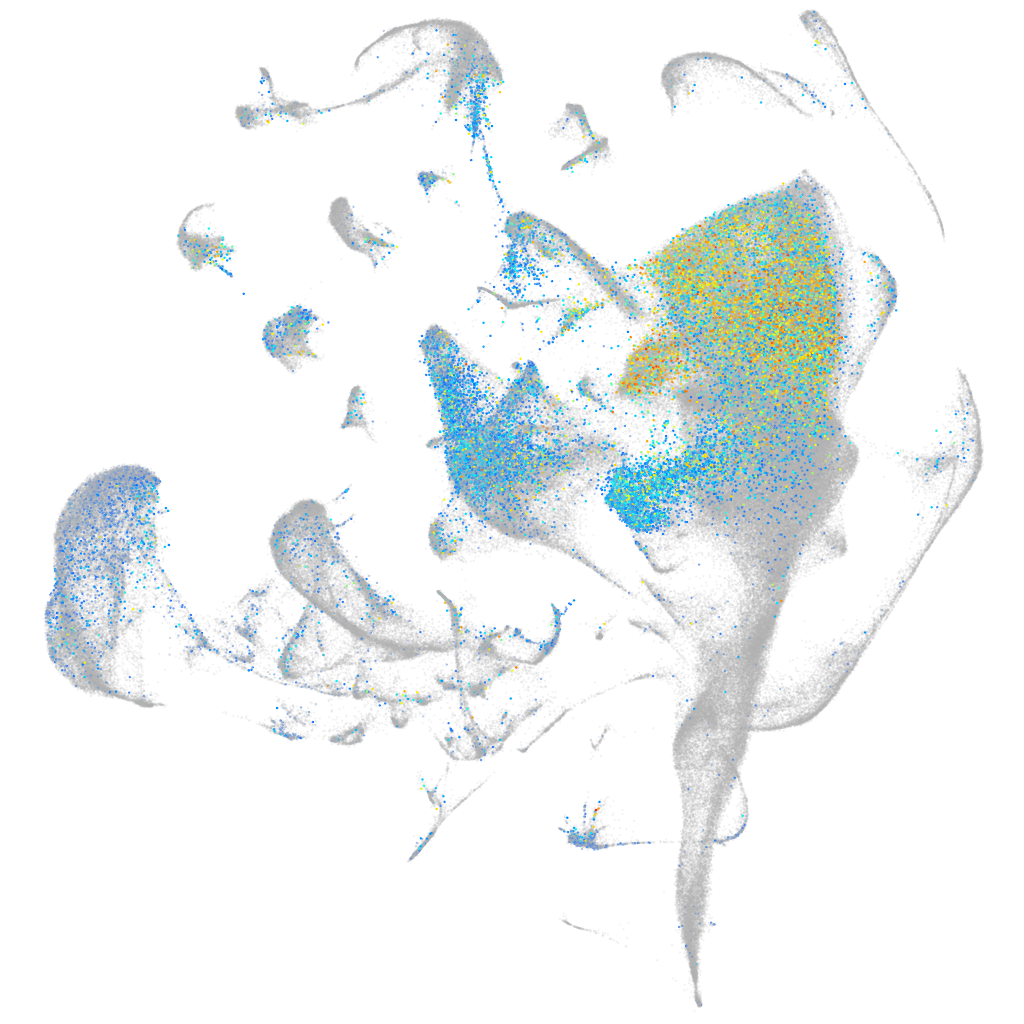nuclear factor I/Xb
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC103909284 | 0.565 | hspb1 | -0.080 |
| nfia | 0.436 | npm1a | -0.076 |
| neurod2 | 0.304 | dkc1 | -0.073 |
| zfpm2a | 0.230 | banf1 | -0.071 |
| neurod6b | 0.227 | eif4ebp3l | -0.071 |
| CU467822.1 | 0.218 | apoeb | -0.070 |
| neurod6a | 0.209 | fbl | -0.070 |
| tcf4 | 0.204 | aldob | -0.068 |
| znf536 | 0.198 | nop58 | -0.068 |
| neurod1 | 0.177 | si:ch73-386h18.1 | -0.068 |
| satb1b | 0.175 | eif5a2 | -0.067 |
| CR848047.1 | 0.174 | lig1 | -0.066 |
| zfpm2b | 0.167 | mki67 | -0.065 |
| stmn1b | 0.161 | zgc:56699 | -0.065 |
| tal1 | 0.161 | chaf1a | -0.064 |
| mdkb | 0.157 | si:ch211-152c2.3 | -0.064 |
| tal2 | 0.157 | zgc:110425 | -0.064 |
| gpm6aa | 0.155 | nhp2 | -0.062 |
| celf2 | 0.154 | rsl1d1 | -0.062 |
| myt1la | 0.146 | zfhx3 | -0.062 |
| ckbb | 0.145 | alcamb | -0.061 |
| tuba1c | 0.144 | gar1 | -0.061 |
| zbtb18 | 0.141 | pcna | -0.061 |
| bhlhe22 | 0.138 | polr3gla | -0.061 |
| mbd3b | 0.133 | s100a1 | -0.061 |
| nova2 | 0.132 | akap12b | -0.060 |
| crabp1a | 0.131 | apoc1 | -0.060 |
| rtn1a | 0.131 | dek | -0.060 |
| id2b | 0.130 | epcam | -0.060 |
| gatad2b | 0.128 | nop56 | -0.060 |
| fez1 | 0.127 | pou5f3 | -0.060 |
| lmo1 | 0.127 | ccnd1 | -0.059 |
| hmgb3a | 0.126 | cdh1 | -0.059 |
| elavl3 | 0.125 | f11r.1 | -0.059 |
| sox4a | 0.124 | stm | -0.059 |