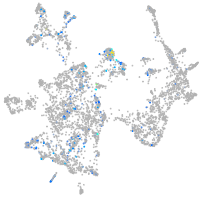
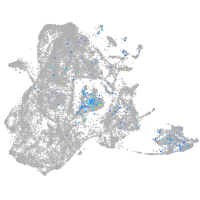
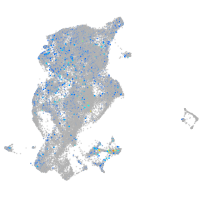
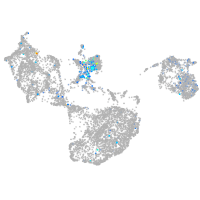
nuclear factor I/Xb
ZFIN 
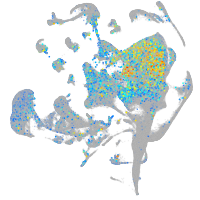
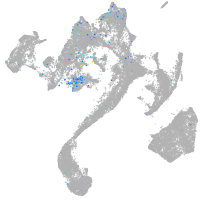
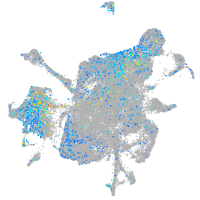
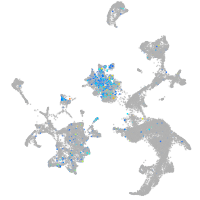
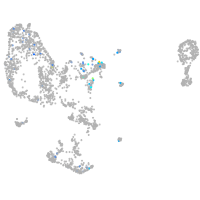
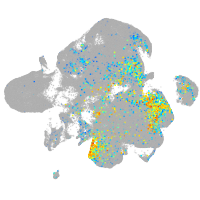
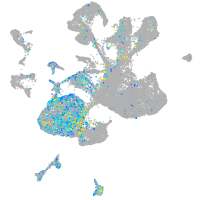
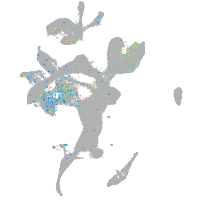
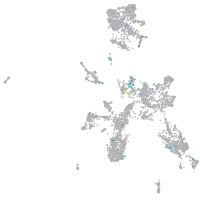
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC103909284 | 0.302 | podxl | -0.065 |
| neurod1 | 0.201 | tmem88b | -0.062 |
| CU467822.1 | 0.199 | rpl35a | -0.059 |
| elavl3 | 0.196 | jam2b | -0.059 |
| nova2 | 0.187 | COLEC10 | -0.059 |
| neurod2 | 0.185 | cd151 | -0.058 |
| CU634008.1 | 0.183 | rpl37 | -0.057 |
| stmn1b | 0.180 | colec11 | -0.057 |
| myt1a | 0.177 | rps19 | -0.056 |
| scrt1a | 0.167 | myrf | -0.055 |
| myt1la | 0.166 | fstl1a | -0.054 |
| rtn1a | 0.159 | metrnlb | -0.053 |
| nfia | 0.157 | fabp11a | -0.053 |
| tuba1c | 0.155 | jcada | -0.053 |
| gng3 | 0.153 | bmp5 | -0.052 |
| si:ch211-137a8.4 | 0.152 | tmem88a | -0.052 |
| adam22 | 0.150 | pkp3b | -0.051 |
| celf3a | 0.146 | si:ch211-286o17.1 | -0.051 |
| si:ch211-1a19.3 | 0.142 | paplna | -0.051 |
| CU469584.1 | 0.142 | ppfibp1a | -0.050 |
| si:ch211-223m11.2 | 0.142 | ecm1b | -0.050 |
| carmil2 | 0.140 | rps12 | -0.050 |
| gpm6aa | 0.138 | phactr4a | -0.049 |
| zfpm2a | 0.138 | prdx1 | -0.049 |
| loxa | 0.136 | cpn1 | -0.049 |
| en2b | 0.134 | si:dkey-195m11.8 | -0.048 |
| gpm6ab | 0.133 | rpl18a | -0.048 |
| zgc:171844 | 0.132 | tinagl1 | -0.048 |
| rtn1b | 0.132 | zbtb8os | -0.048 |
| cdk5r1b | 0.131 | col7a1l | -0.048 |
| si:ch211-146m13.3 | 0.129 | vegfaa | -0.047 |
| ebf3a | 0.129 | LOC101884451 | -0.047 |
| sox14 | 0.128 | klf6a | -0.046 |
| tal1 | 0.127 | aldh9a1a.1 | -0.046 |
| hmgb3a | 0.126 | angptl6 | -0.045 |