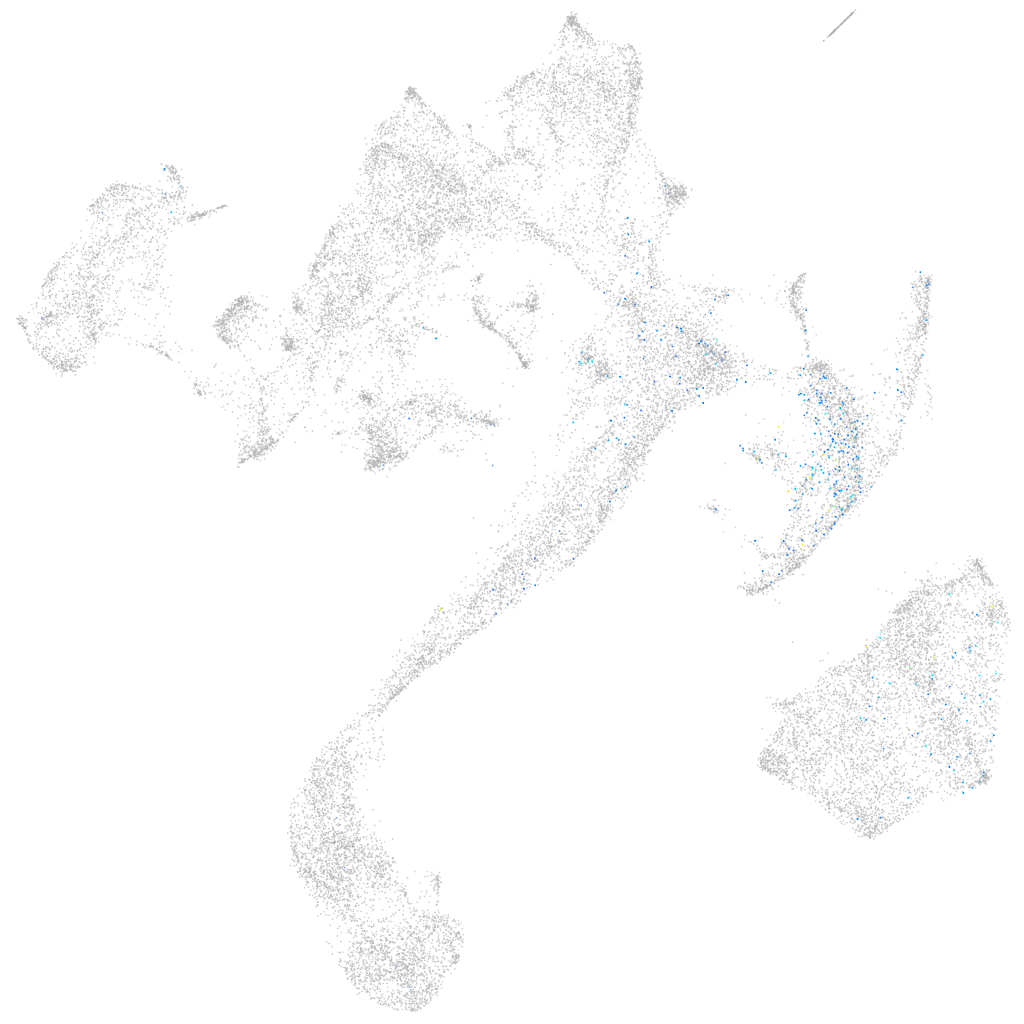
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ece1 | 0.156 | fabp3 | -0.079 |
| her1 | 0.150 | COX3 | -0.070 |
| hoxd10a | 0.133 | bhmt | -0.068 |
| msgn1 | 0.131 | actc1b | -0.064 |
| XLOC-039166 | 0.128 | sparc | -0.063 |
| apoc1 | 0.123 | mt-co2 | -0.063 |
| tbx16l | 0.122 | eef1da | -0.059 |
| itm2cb | 0.122 | gamt | -0.059 |
| her7 | 0.120 | ckma | -0.058 |
| LOC110439323 | 0.119 | col1a2 | -0.058 |
| hoxb10a | 0.116 | atp5l | -0.057 |
| pcdh8 | 0.116 | ckmb | -0.057 |
| BX511166.1 | 0.115 | ttn.2 | -0.057 |
| dgkb | 0.115 | pabpc4 | -0.056 |
| BX005254.3 | 0.115 | atp2a1 | -0.055 |
| fn1b | 0.113 | col1a1a | -0.055 |
| hes6 | 0.106 | pmp22a | -0.054 |
| her12 | 0.105 | celf2 | -0.054 |
| dlc | 0.104 | ak1 | -0.053 |
| nid2a | 0.104 | pvalb1 | -0.053 |
| igsf9a | 0.099 | ndrg2 | -0.053 |
| ripply2 | 0.097 | si:dkey-16p21.8 | -0.053 |
| cx43.4 | 0.097 | mylpfa | -0.053 |
| myf5 | 0.095 | eno1a | -0.052 |
| apoeb | 0.094 | pvalb2 | -0.052 |
| LOC110438146 | 0.093 | aldoab | -0.052 |
| pcmtd1 | 0.092 | vwde | -0.051 |
| efnb2b | 0.092 | tmem38a | -0.051 |
| gnaia | 0.092 | cd81a | -0.051 |
| draxin | 0.091 | tnnt3a | -0.050 |
| tbx6 | 0.091 | hbbe1.3 | -0.050 |
| hoxd11a | 0.089 | neb | -0.050 |
| XLOC-042934 | 0.089 | gapdh | -0.050 |
| LO016987.2 | 0.088 | si:ch211-140m22.7 | -0.050 |
| fbln1 | 0.087 | foxp4 | -0.050 |