grainyhead-like transcription factor 2b
ZFIN 
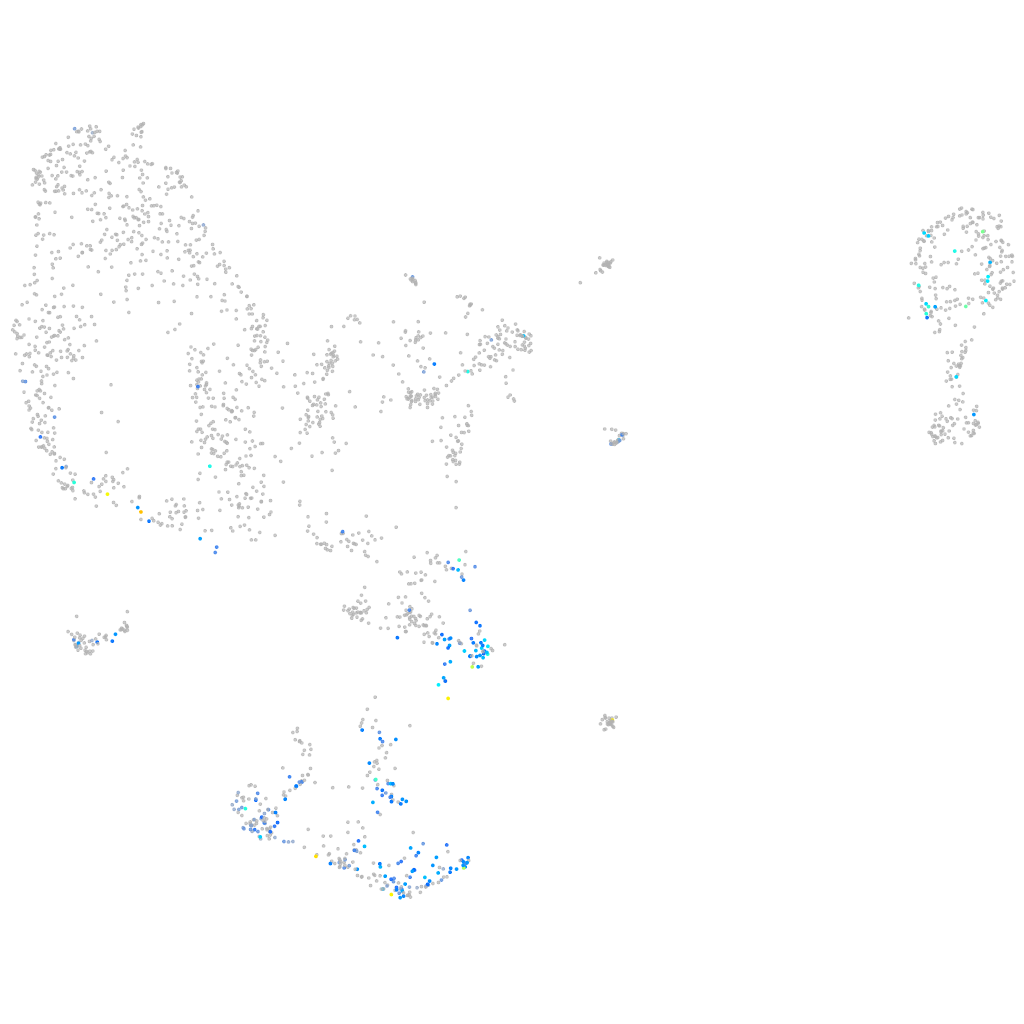
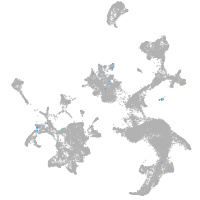
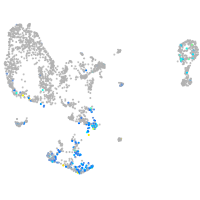
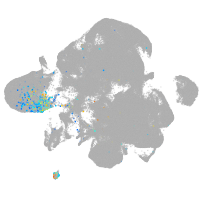
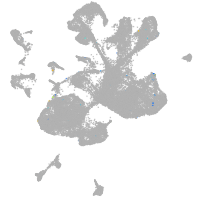
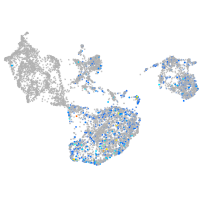
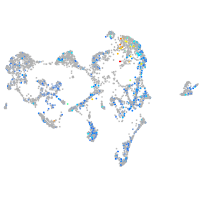
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tfap2a | 0.353 | dab2 | -0.226 |
| hoxc11b | 0.333 | grhprb | -0.223 |
| cldnh | 0.304 | si:dkey-194e6.1 | -0.221 |
| tbx2a | 0.289 | aqp8b | -0.211 |
| phlda2 | 0.284 | zgc:101040 | -0.207 |
| hoxb10a | 0.282 | lrp2a | -0.204 |
| gata3 | 0.279 | fbp1b | -0.203 |
| dnase1l4.1 | 0.278 | pdzk1ip1 | -0.202 |
| rnf183 | 0.275 | nit2 | -0.199 |
| foxa3 | 0.266 | ahcyl1 | -0.197 |
| zgc:158423 | 0.260 | aldob | -0.197 |
| mylk5 | 0.258 | si:dkey-28n18.9 | -0.196 |
| si:ch211-153b23.5 | 0.255 | ftcd | -0.196 |
| si:dkey-36i7.3 | 0.253 | zgc:136493 | -0.196 |
| degs2 | 0.253 | pnp6 | -0.195 |
| atp1a1a.3 | 0.252 | slc26a1 | -0.195 |
| mal2 | 0.252 | upb1 | -0.195 |
| mapkapk3 | 0.249 | gstt1a | -0.194 |
| ftr83 | 0.248 | cubn | -0.194 |
| hoxd12a | 0.248 | hao1 | -0.192 |
| trim35-12 | 0.247 | slc9a3r1a | -0.190 |
| tfap2b | 0.247 | gamt | -0.189 |
| bsnd | 0.246 | si:dkey-33i11.9 | -0.188 |
| isl2a | 0.245 | alpl | -0.187 |
| clcnk | 0.237 | pdzk1 | -0.186 |
| cldne | 0.237 | gpt2l | -0.186 |
| etf1a | 0.237 | dpys | -0.183 |
| tmem176l.4 | 0.237 | pklr | -0.180 |
| soul2 | 0.236 | gstr | -0.179 |
| hoxb8a | 0.234 | lrpap1 | -0.174 |
| glsb | 0.233 | sord | -0.174 |
| ppp1r1b | 0.232 | slc4a4a | -0.173 |
| f11r.1 | 0.231 | ASS1 | -0.173 |
| hoxb9a | 0.230 | agxta | -0.173 |
| si:ch73-359m17.9 | 0.228 | suclg2 | -0.173 |