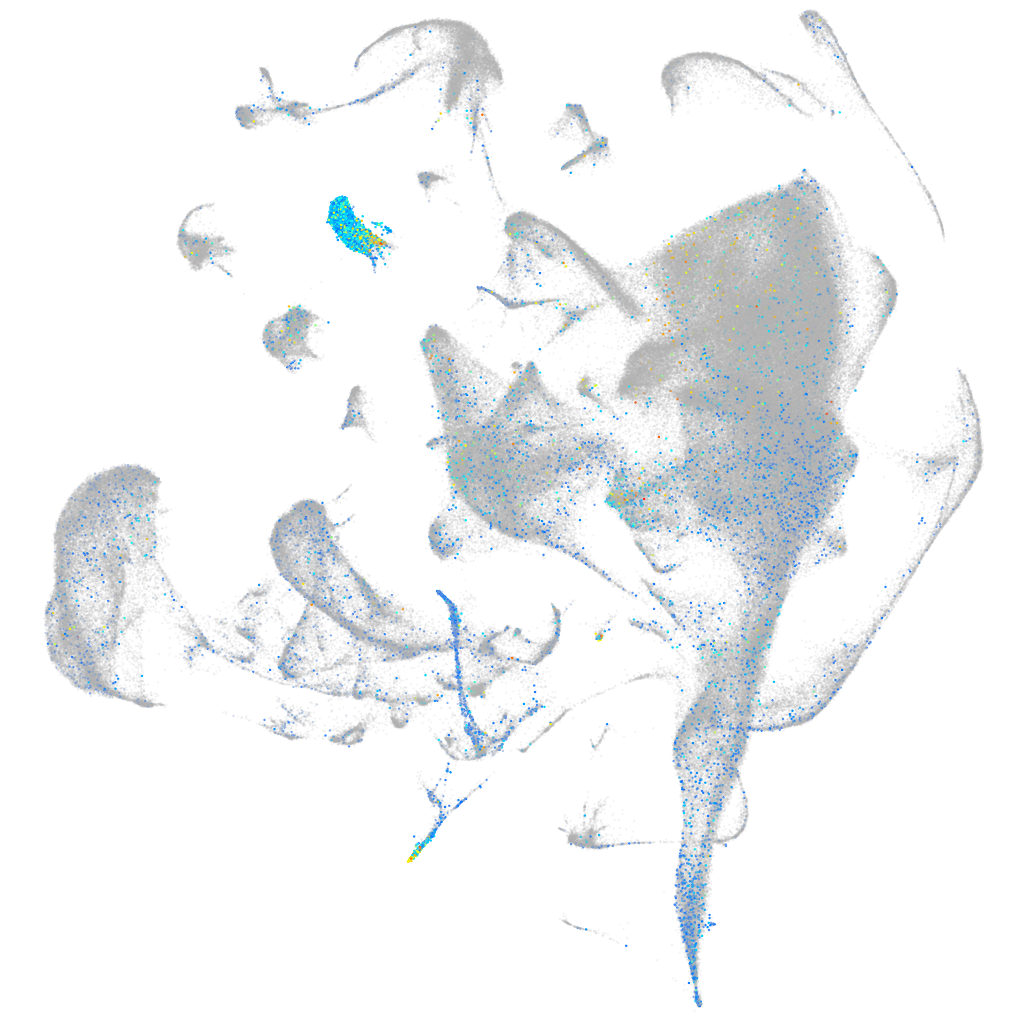sorbitol dehydrogenase
ZFIN 
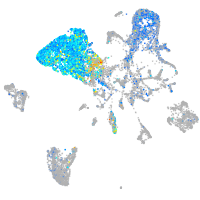
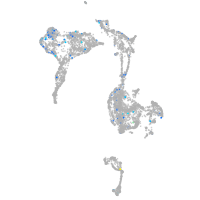
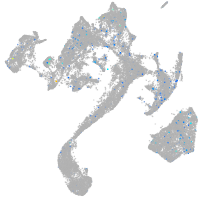
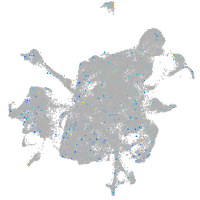
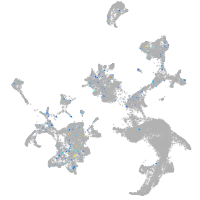
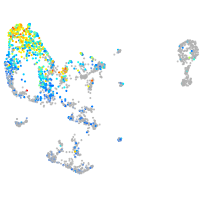
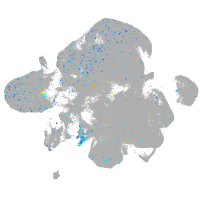
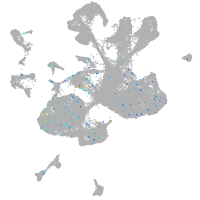
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hao1 | 0.609 | tuba1c | -0.089 |
| grhprb | 0.603 | marcksl1a | -0.087 |
| agxta | 0.583 | hmgb3a | -0.084 |
| f2 | 0.577 | nova2 | -0.084 |
| ftcd | 0.577 | gpm6aa | -0.082 |
| agxtb | 0.571 | hmgb1b | -0.081 |
| c9 | 0.560 | rtn1a | -0.080 |
| serpinf2a | 0.560 | atp6v0cb | -0.078 |
| dpys | 0.558 | jpt1b | -0.078 |
| cfhl4 | 0.556 | elavl3 | -0.077 |
| hpda | 0.556 | gpm6ab | -0.077 |
| shbg | 0.555 | tubb5 | -0.075 |
| cfb | 0.554 | hnrnpa0a | -0.074 |
| si:ch211-288g17.4 | 0.554 | tuba1a | -0.073 |
| fbp1b | 0.553 | rnasekb | -0.071 |
| g6pca.2 | 0.553 | ckbb | -0.070 |
| hal | 0.553 | stmn1b | -0.070 |
| zgc:112265 | 0.553 | tmeff1b | -0.070 |
| gc | 0.552 | celf2 | -0.068 |
| serpina1 | 0.552 | si:ch1073-429i10.3.1 | -0.067 |
| fga | 0.549 | fam168a | -0.066 |
| sec14l7 | 0.549 | gapdhs | -0.066 |
| tymp | 0.549 | zc4h2 | -0.066 |
| fgb | 0.548 | marcksb | -0.065 |
| serpina1l | 0.548 | mdkb | -0.065 |
| si:dkey-86l18.10 | 0.548 | rtn1b | -0.065 |
| f7i | 0.547 | si:dkey-276j7.1 | -0.065 |
| serpinf2b | 0.547 | chd4a | -0.064 |
| a2ml | 0.546 | epb41a | -0.063 |
| fgg | 0.546 | sncb | -0.063 |
| ambp | 0.545 | cadm3 | -0.062 |
| pklr | 0.544 | tmsb | -0.062 |
| apom | 0.543 | vamp2 | -0.062 |
| urad | 0.543 | atpv0e2 | -0.061 |
| kng1 | 0.541 | gng3 | -0.061 |