crestin
ZFIN 
Other cell groups


all cells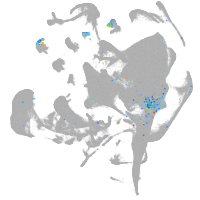 |
blastomeres |
PGCs |
endoderm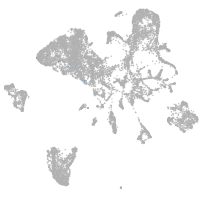 |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle 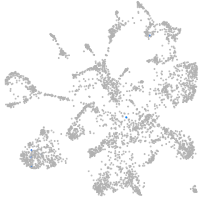 |
mesenchyme 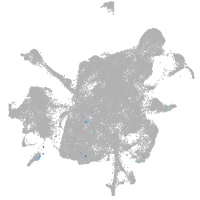 |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ednrab | 0.785 | si:ch73-1a9.3 | -0.038 |
| LOC110438542 | 0.472 | tuba1c | -0.038 |
| BX248318.1 | 0.444 | gpm6aa | -0.037 |
| si:ch211-11c3.9 | 0.411 | cadm3 | -0.035 |
| si:dkey-19a16.13 | 0.378 | ckbb | -0.034 |
| si:dkey-250k10.4 | 0.372 | si:ch211-137a8.4 | -0.032 |
| si:ch211-11c3.12 | 0.352 | epb41a | -0.031 |
| XLOC-001964 | 0.332 | ptmab | -0.031 |
| LOC108179355 | 0.322 | fabp7a | -0.030 |
| sox10 | 0.300 | pvalb1 | -0.029 |
| LOC110439533 | 0.296 | nova2 | -0.029 |
| slc15a2 | 0.295 | rnasekb | -0.028 |
| msx1b | 0.293 | hmgb1a | -0.028 |
| erbb3b | 0.291 | ptmaa | -0.028 |
| inka1a | 0.278 | COX3 | -0.027 |
| olfcg4 | 0.264 | pvalb2 | -0.027 |
| hoxb7a | 0.263 | anp32e | -0.027 |
| hoxc3a | 0.261 | foxg1b | -0.025 |
| slc1a3a | 0.254 | snap25b | -0.025 |
| BX005254.3 | 0.221 | gpm6ab | -0.025 |
| mov10b.1 | 0.221 | gapdhs | -0.024 |
| tfec | 0.221 | atp6v0cb | -0.024 |
| BX001014.2 | 0.216 | hmgb1b | -0.023 |
| ets1 | 0.214 | rorb | -0.023 |
| eva1ba | 0.210 | mt-co2 | -0.022 |
| FO704721.1 | 0.204 | sypb | -0.021 |
| XLOC-042222 | 0.190 | hbbe1.3 | -0.021 |
| pax3a | 0.188 | actc1b | -0.021 |
| epoa | 0.185 | syt5b | -0.021 |
| SCARF2 | 0.183 | otx5 | -0.021 |
| tes | 0.175 | atpv0e2 | -0.021 |
| XLOC-013009 | 0.174 | ndrg4 | -0.021 |
| crlf1a | 0.171 | tmpob | -0.020 |
| si:ch73-156l6.2 | 0.171 | neurod4 | -0.020 |
| BX927308.2 | 0.168 | tubb2b | -0.020 |




