crestin
ZFIN 
Other cell groups


all cells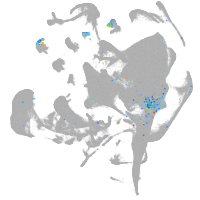 |
blastomeres |
PGCs |
endoderm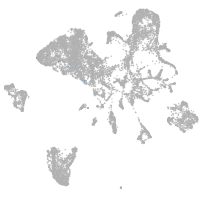 |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle 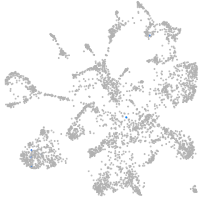 |
mesenchyme 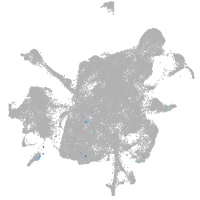 |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ednrab | 0.648 | nova2 | -0.069 |
| tfec | 0.598 | tuba1c | -0.064 |
| snai1b | 0.339 | gpm6aa | -0.060 |
| si:dkey-19a16.13 | 0.335 | ckbb | -0.048 |
| LOC110438542 | 0.335 | rtn1a | -0.046 |
| si:ch211-11c3.9 | 0.325 | fabp7a | -0.046 |
| LOC110439533 | 0.288 | elavl3 | -0.045 |
| BX248318.1 | 0.284 | rnasekb | -0.044 |
| si:ch211-11c3.12 | 0.270 | pvalb2 | -0.043 |
| inka1a | 0.259 | tmsb | -0.042 |
| trpm1b | 0.238 | ptmaa | -0.042 |
| slc15a2 | 0.234 | sox3 | -0.041 |
| znf1004 | 0.220 | pvalb1 | -0.041 |
| crlf1a | 0.218 | CU467822.1 | -0.040 |
| ets1 | 0.202 | mdka | -0.040 |
| mov10b.1 | 0.201 | ncam1a | -0.040 |
| sfrp2 | 0.196 | vim | -0.040 |
| dlc1 | 0.195 | si:dkey-56m19.5 | -0.039 |
| erbb3b | 0.192 | zgc:165461 | -0.038 |
| itga9 | 0.187 | hbbe1.3 | -0.038 |
| si:dkey-250k10.4 | 0.164 | gpm6ab | -0.038 |
| pdgfra | 0.163 | si:ch211-137a8.4 | -0.038 |
| msx1b | 0.159 | actc1b | -0.038 |
| sox10 | 0.158 | stmn1b | -0.037 |
| XLOC-001603 | 0.157 | tox | -0.037 |
| tfap2a | 0.151 | slc1a2b | -0.036 |
| cyth1b | 0.147 | cadm3 | -0.036 |
| LOC108179355 | 0.146 | epb41a | -0.035 |
| tspan36 | 0.146 | CU634008.1 | -0.035 |
| tmem119b | 0.144 | hmgb1b | -0.035 |
| XLOC-042222 | 0.143 | msi1 | -0.035 |
| msx2b | 0.141 | dpysl2b | -0.035 |
| SCARF2 | 0.138 | nr2f1a | -0.034 |
| CABZ01088025.1 | 0.138 | cdh2 | -0.034 |
| eva1ba | 0.136 | gapdhs | -0.034 |




