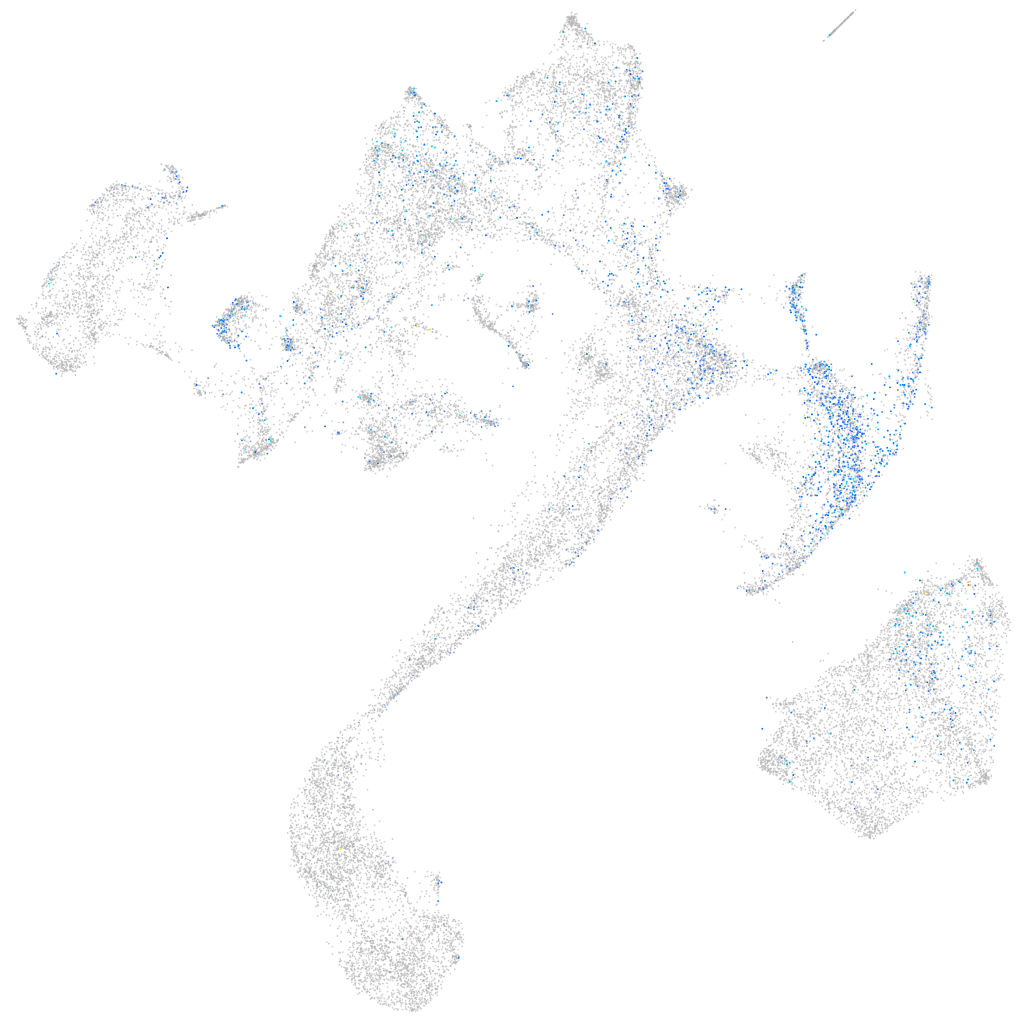
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| her1 | 0.218 | actc1b | -0.112 |
| pcdh8 | 0.211 | ckmb | -0.112 |
| nid2a | 0.176 | atp2a1 | -0.111 |
| fn1b | 0.172 | ckma | -0.110 |
| her7 | 0.171 | gapdh | -0.108 |
| itm2cb | 0.168 | pabpc4 | -0.107 |
| tbx6 | 0.168 | ak1 | -0.107 |
| tspan7 | 0.166 | aldoab | -0.101 |
| myf5 | 0.166 | tnnc2 | -0.101 |
| ephb3 | 0.162 | mylpfa | -0.101 |
| msgn1 | 0.160 | ldb3b | -0.100 |
| hoxb10a | 0.157 | eno1a | -0.099 |
| draxin | 0.157 | gamt | -0.099 |
| her12 | 0.156 | nme2b.2 | -0.098 |
| hoxd10a | 0.151 | tmem38a | -0.098 |
| phc2a | 0.150 | acta1b | -0.098 |
| ripply2 | 0.148 | si:dkey-16p21.8 | -0.097 |
| apoc1 | 0.148 | si:ch73-367p23.2 | -0.097 |
| BX005254.3 | 0.146 | ank1a | -0.096 |
| LO016987.2 | 0.145 | neb | -0.096 |
| XLOC-042222 | 0.144 | actn3a | -0.096 |
| gnaia | 0.143 | pvalb1 | -0.096 |
| has2 | 0.142 | mylz3 | -0.095 |
| dlc | 0.139 | myl1 | -0.095 |
| her11 | 0.139 | myom1a | -0.095 |
| tbx16l | 0.137 | pvalb2 | -0.094 |
| sall4 | 0.136 | tnnt3a | -0.094 |
| hnrnpa0l | 0.135 | pgam2 | -0.094 |
| cdh2 | 0.133 | smyd1a | -0.093 |
| cx43.4 | 0.133 | hhatla | -0.093 |
| ism1 | 0.133 | atp1a2a | -0.092 |
| metrnla | 0.130 | ldb3a | -0.092 |
| fn1a | 0.129 | tmod4 | -0.092 |
| ubc | 0.129 | CABZ01078594.1 | -0.091 |
| alpi.1 | 0.128 | srl | -0.091 |