protein tyrosine phosphatase 4A2b
ZFIN 
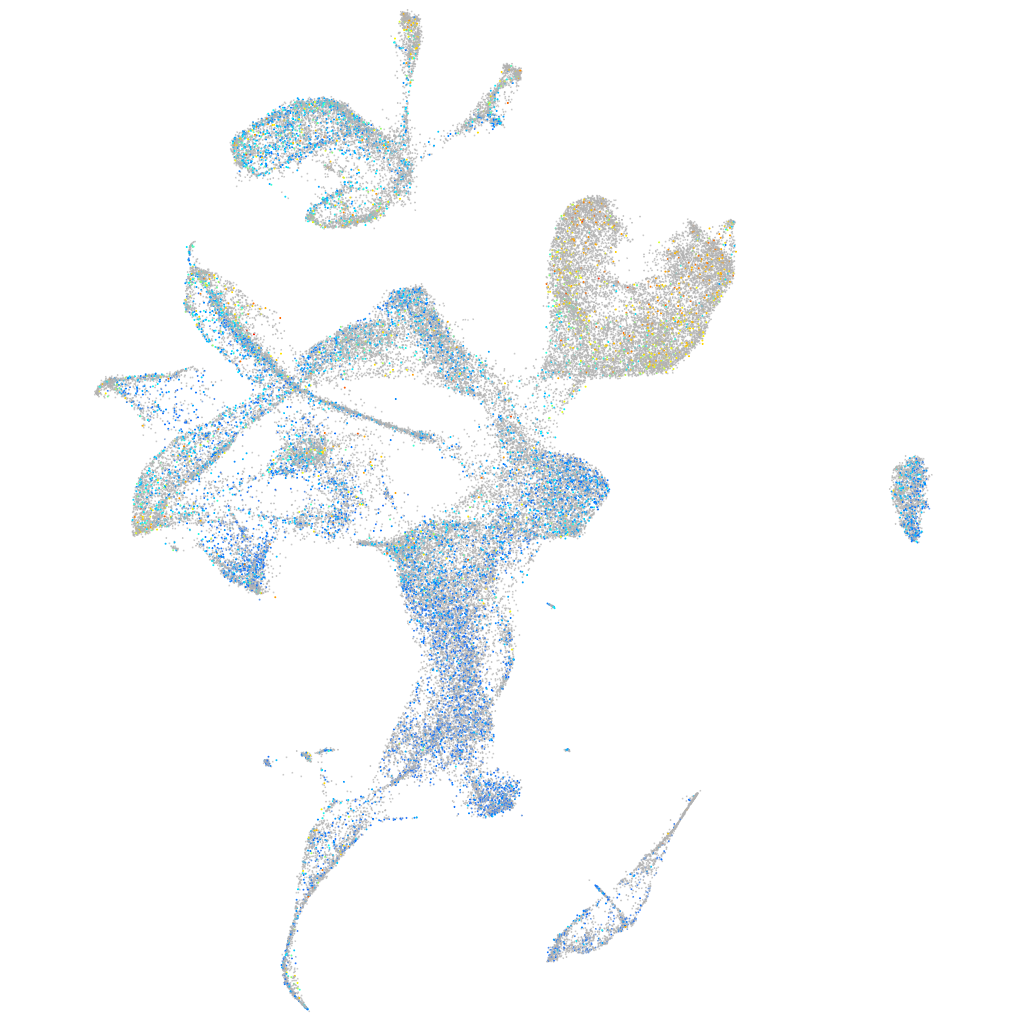
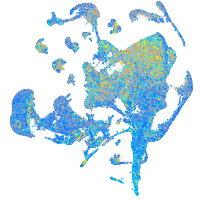
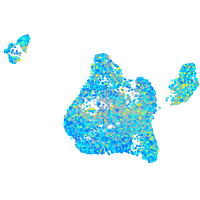
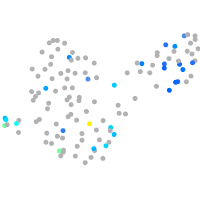
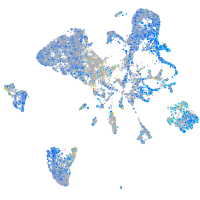
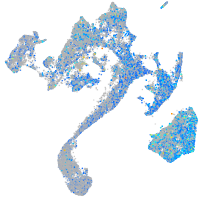
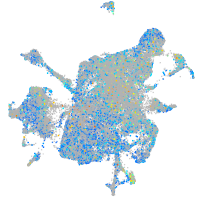
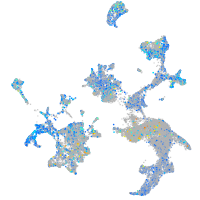
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sncb | 0.079 | actc1b | -0.035 |
| rtn1b | 0.071 | cryba4 | -0.031 |
| tuba1c | 0.070 | cryba1l1 | -0.030 |
| stx1b | 0.068 | cryba1b | -0.030 |
| ppp1r14ba | 0.068 | crybb1l2 | -0.029 |
| eno2 | 0.068 | cryba2a | -0.029 |
| vamp2 | 0.068 | lim2.4 | -0.029 |
| zgc:153426 | 0.067 | cryba2b | -0.029 |
| ywhag2 | 0.066 | crybb1l1 | -0.028 |
| stxbp1a | 0.065 | crygn2 | -0.028 |
| zgc:65894 | 0.065 | fabp11a | -0.028 |
| si:ch73-290k24.5 | 0.063 | cryba1l2 | -0.028 |
| gnb1a | 0.063 | crybb1 | -0.028 |
| snap25a | 0.062 | crygmx | -0.027 |
| celf5a | 0.061 | cx23 | -0.026 |
| stmn2a | 0.061 | pvalb2 | -0.026 |
| elavl4 | 0.059 | lim2.1 | -0.026 |
| elavl3 | 0.058 | BFSP1 | -0.025 |
| gnb1b | 0.058 | gpx9 | -0.025 |
| tuba2 | 0.058 | lim2.3 | -0.025 |
| ptp4a1 | 0.057 | mipa | -0.025 |
| cplx2l | 0.057 | crygm2d13 | -0.025 |
| gng3 | 0.057 | si:ch211-255g12.6 | -0.024 |
| gnao1a | 0.057 | crygm2d10 | -0.024 |
| syt1a | 0.056 | crygm2d8 | -0.024 |
| maptb | 0.056 | crybb1l3 | -0.023 |
| syn2a | 0.056 | mipb | -0.023 |
| sv2a | 0.056 | gja8b | -0.023 |
| ywhabl | 0.054 | si:ch211-214j24.14 | -0.023 |
| sprn | 0.054 | tubb6 | -0.023 |
| ywhaz | 0.054 | BX545855.1 | -0.022 |
| gabrb4 | 0.054 | pvalb1 | -0.022 |
| rab2a | 0.053 | crygm2d12 | -0.022 |
| rbfox1 | 0.053 | prox2 | -0.022 |
| calm2b | 0.053 | crygmxl2 | -0.021 |