"crystallin, gamma MX, like 2"
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye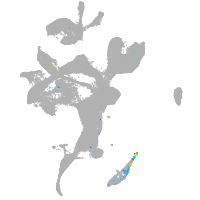 |
taste / olfactory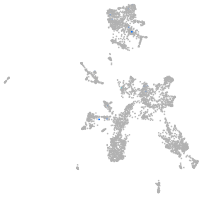 |
otic / lateral line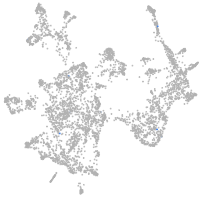 |
epidermis |
periderm |
fin |
ionocytes / mucous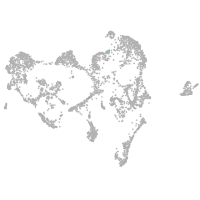 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cryba1l2 | 0.362 | aldob | -0.017 |
| capn3a | 0.361 | ctsla | -0.017 |
| BX548032.3 | 0.348 | h2afva | -0.016 |
| crygm2d10 | 0.338 | mcl1a | -0.015 |
| BX545855.1 | 0.333 | atp5if1a | -0.014 |
| crygm2d12 | 0.331 | ier2b | -0.014 |
| lim2.4 | 0.330 | adh5 | -0.013 |
| crygm2d13 | 0.330 | anp32a | -0.013 |
| crygm2d5 | 0.330 | dbi | -0.013 |
| crygm2d21 | 0.324 | fosab | -0.013 |
| crygm2d18 | 0.322 | fxyd6l | -0.013 |
| crygm2d4 | 0.321 | nucks1a | -0.013 |
| crybb1l1 | 0.320 | sod1 | -0.013 |
| crygm2d7 | 0.318 | zgc:162730 | -0.013 |
| crybb1l2 | 0.314 | anp32e | -0.012 |
| crygm2d3 | 0.311 | cbx5 | -0.012 |
| mipa | 0.308 | ccni | -0.012 |
| cryba2a | 0.307 | cd63 | -0.012 |
| cx23 | 0.307 | cebpd | -0.012 |
| crygm2d8 | 0.306 | chaf1a | -0.012 |
| crygm2d20 | 0.303 | cx43.4 | -0.012 |
| lim2.3 | 0.303 | jun | -0.012 |
| cryba1b | 0.301 | marcksb | -0.012 |
| cryba1l1 | 0.295 | mcl1b | -0.012 |
| lim2.1 | 0.294 | nasp | -0.012 |
| crygm2d1 | 0.289 | si:ch211-196l7.4 | -0.012 |
| endou2 | 0.288 | si:ch211-288g17.3 | -0.012 |
| crygmx | 0.287 | ucp2 | -0.012 |
| crybb1 | 0.286 | aldh9a1a.1 | -0.011 |
| gpx9 | 0.283 | btg1 | -0.011 |
| crygm2d2 | 0.282 | capns1a | -0.011 |
| crygn2 | 0.276 | cbx3a | -0.011 |
| lim2.5 | 0.275 | cdkn1a | -0.011 |
| cryba4 | 0.270 | cebpb | -0.011 |
| BFSP1 | 0.268 | ddit3 | -0.011 |




