glutathione peroxidase 9
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 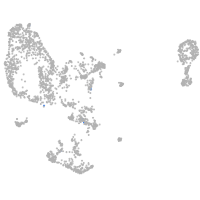 |
neural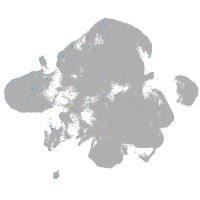 |
spinal cord / glia |
eye |
taste / olfactory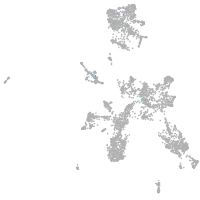 |
otic / lateral line |
epidermis |
periderm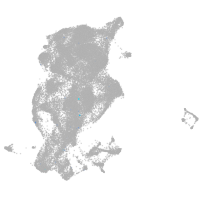 |
fin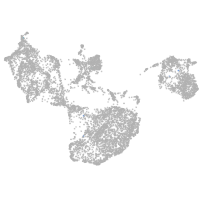 |
ionocytes / mucous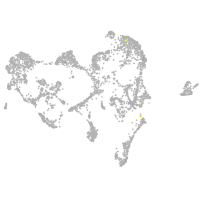 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cryba1l2 | 0.382 | fabp3 | -0.019 |
| lim2.4 | 0.354 | fabp7a | -0.018 |
| mipa | 0.351 | adh5 | -0.017 |
| cx23 | 0.343 | gpm6aa | -0.017 |
| BX548032.3 | 0.339 | hmgb1b | -0.017 |
| capn3a | 0.338 | sod1 | -0.017 |
| crybb1l1 | 0.337 | krt4 | -0.015 |
| cryba2a | 0.332 | mcl1a | -0.015 |
| lim2.3 | 0.331 | mdkb | -0.015 |
| crybb1l2 | 0.330 | zgc:162730 | -0.015 |
| lim2.5 | 0.328 | anxa1a | -0.014 |
| cryba1b | 0.314 | cebpd | -0.014 |
| lim2.1 | 0.310 | ckbb | -0.014 |
| cryba1l1 | 0.309 | ctsla | -0.014 |
| mipb | 0.307 | CU467822.1 | -0.014 |
| crybb1l3 | 0.304 | fxyd6l | -0.014 |
| crygmx | 0.304 | h2afva | -0.014 |
| gja8b | 0.299 | CU634008.1 | -0.014 |
| crybb1 | 0.294 | ccni | -0.013 |
| crygn2 | 0.290 | cd63 | -0.013 |
| crygmxl2 | 0.283 | cebpb | -0.013 |
| BFSP1 | 0.281 | cyt1 | -0.013 |
| gja8a | 0.278 | krtt1c19e | -0.013 |
| cryba4 | 0.277 | mgst1.2 | -0.013 |
| endou2 | 0.276 | prdx2 | -0.013 |
| cryba2b | 0.272 | pycard | -0.013 |
| grifin | 0.267 | si:ch211-137a8.4 | -0.013 |
| crygm2d10 | 0.265 | tuba1c | -0.013 |
| crygm2d13 | 0.263 | dap1b | -0.013 |
| si:ch211-255g12.6 | 0.254 | aldh9a1a.1 | -0.012 |
| bfsp2 | 0.247 | celf2 | -0.012 |
| crygm2d8 | 0.246 | cfl1l | -0.012 |
| cps1 | 0.245 | chd4a | -0.012 |
| crygm2d12 | 0.245 | gpm6ab | -0.012 |
| BX545855.1 | 0.241 | ier2b | -0.012 |




